The IVIVC object provides an organized environment in which to build and manipulate InVitro-InVivo Correlation (IVIVC) models. The IVIVC object also estimates the unit impulse response (UIR) for one or more profiles given an IV (bolus), infusion, or oral formulation. The IVIVC object supports specification and running of all or a subset of operations required to create an IVIVC model, validate it, and use it to make PK predictions.
Note that InVitro and InVivo data must be in separate datasets. Individual functions are specified and run in the separate panels and tabs of the IVIVC object.
The IVIVC object contains several sub-workflows or steps. These steps can be executed individually during the IVIVC modeling, i.e., it is not necessary to execute the entire object during the IVIVC modeling process. When an IVIVC object is executed, however, all IVIVC steps will run, even if the user only wants to run one step.
Note:If the data for a step is fed into the workflow (e.g., through a datalink), executing the IVIVC step from the step’s properties panel will not execute the source feeder and can lead to a “lack of source” error. If this occurs, execute the full IVIVC workflow object to ensure that sources are executed prior to the IVIVC workflow validation.
In vitro data, in vivo data, and dosing data should be similarly scaled for a successful IVIVC (see the “IVIVC workflow example” for an example of a full IVIVC workflow). Phoenix IVIVC uses a two-stage Level A correlation, where deconvolution is followed by a linear correlation model representing a relationship between in vitro dissolution and in vivo input rate. The in vitro dissolution and in vivo input curves should be directly superimposable or be made superimposable by using a scaling factor (FDA 1997 Guidance on IVIVC) or by changing units to convert data appropriately. For example, specifying in vivo dosing as a very large value will result in large input rates from deconvolution. If dissolution data is then given in small-scale numbers, Level A IVIVC may fail to produce accurate results due to numerical instabilities, particularly Convolution in the prediction stage may fail to produce non-zero concentrations. The dosing data in this case should be scaled by using larger units (e.g., convert ng to mg) in order for the input rates to be consistently scaled with the dissolution data.
Use one of the following to add a IVIVC object to a Workflow:
Right-click menu for a Workflow object: New > IVIVC > IVIVC.
Main menu: Insert > IVIVC > IVIVC.
Right-click menu for a worksheet: Send To > IVIVC > IVIVC.
Note:To view the object in its own window, select it in the Object Browser and double-click it or press ENTER. All instructions for setting up and execution are the same whether the object is viewed in its own window or in Phoenix view.
This section contains the following topics:
-
InVitro panels and tab
-
InVivo panels and tab
-
Prediction panels and tab
The IVIVC object Status panel displays the status of steps that can be completed in the process of creating, testing, and using the IVIVC object. Each part of the IVIVC process is listed beside a square that uses colors to denote the status of that process.
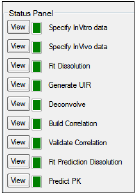
Click View beside the different steps in the Status Panel to view the associated panel, tab, or dataset. This also filters the Results tab list to those generated by the selected operation.
Note:It is not necessary to complete every step to run an IVIVC model.
Use the Main Mappings panel to identify how input variables are used in a IVIVC object. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•InVitro Time: Sampling time values. Column type must be numeric.
•InVitro Dissolution: Dissolution data.
•InVitro Formulation: Formulation identifiers.
Note:“Formulation” is a reserved word for InVitro Data. If a column named “Formulation” is mapped to InVitro Formulation, it can prevent any requested Levy Plots from being generated. In such a case, the IVIVC object must be recreated, settings reapplied, the Formulation column’s name changed, remapped, and the object re-executed.
Formulation data that is not mapped in the InVitro Formulation panel is still included in the output plots Correlation overlay, Levy plot, and Fabs vs Fdiss. Use the resulting worksheets and create plots after filtering the formulation that should not be included (e.g., for the Levy plot, filter unwanted data from the “Levy Plots.Tvivo vs Tvitro.Levy Plot Values” worksheet; for the Correlation overlay plot, filter unwanted data from the “Correlation.Abs vs Diss Data” worksheet). In addition, you must specify the data to plot the line of unity.
The InVitro Formulation panel allows users to partition dissolution data. Use the option buttons to assign formulations to either internal or external validation. Formulations marked as internal (there must be at least one chosen) are used to estimate the correlation (IVIVC). Formulations marked as external are only used for validation of the correlation (IVIVC). The InVitro Formulation panel is automatically populated with formulations taken from the dataset mapped to the InVitro Data panel. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Internal: Validation: Formulation(s) used to build the IVIVC model(s) and for validation on the Correlation tab. See “Correlation tab” for more information.
•External: Validation: Formulation(s) used in model validation.
After the InVitro dissolution and formulation data are mapped, users can specify parameter values for different formulations in the InVitro Estimates panel. See “InVitro tab” for initial value, and lower and upper bound options.
Depending on the model, users can fix certain parameters to known values. For example, the fraction dissolved, Finf, can be fixed to 1.0 to prevent incomplete measurements from biasing other parameter estimates.
All iterative estimation procedures require initial estimates of the parameters. Phoenix can compute initial estimates via curve stripping for single-dose PK, PD, PK/PD link, linear or IVIVC models. For all other situations, users must supply initial estimates or provide boundaries to be used by Phoenix in creating initial estimates via grid search.
For IVIVC models only, some parameters can be assigned fixed values while others are estimated. The fixed command can be used for both modeling and simulating. Fixed parameters use a set value, so are no longer considered parameters. This reduces the number of parameters in the model. Statistics such as VIF are not computed for fixed parameters.
•None: Data types mapped to this context are not included in any analysis or output.
•Sort: Map the formulation data to the Sort context to sort the parameter estimates by formulation.
•Parameter: IVIVC model parameters.
•Initial: Initial estimates for the parameter.
•Fixed or Estimated: Whether or not the initial parameter is fixed or estimated.
•Lower: (Available when User Supplied Bounds is selected in the InVitro tab) Lower boundary used for parameter estimates.
•Upper: (Available when User Supplied Bounds is selected in the InVitro tab) Upper boundary used for parameter estimates.
Note:The Fixed or Estimated and Initial columns will be reset to default values if the data source is switched from an external worksheet to an internal worksheet. You will need to make those changes again for the internal worksheet.
In order to provide smoothing, users can fit the dissolution data to a sigmoidal dissolution model.
-
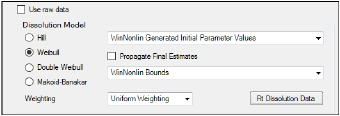
Check the Use raw data checkbox to perform linear interpolation on the raw data instead of modeling.
-
Select an option button in the Dissolution Model area to select a model. Users can select one of four dissolution models: Hill, Weibull, Double Weibull, Makoid-Banakar.
For descriptions of these models, see “Sigmoidal and Dissolution models”. -
In the Weighting menu, select one of five methods for weighting data during modeling (see “Weighting” in the WinNonlin NCA section for explanations of the various schemes).
•Uniform Weighting
•1/Y (weight by 1/observed Y)
•1/Yhat (weight by 1/predicted Y [iterative reweighting])
•1/(Y*Y) (weight by 1/observed Y2)
•1(Yhat*Yhat) (weight by 1/predicted Y2 [iterative reweighting])
-
Specify how to determine initial values:
•Select User Supplied Initial Parameter Values to enter initial parameter estimates in the InVitro Estimates panel.
•Select WinNonlin Generated Initial Parameter Values to have Phoenix determine the initial parameter values.
-
Check the Propagate Final Estimates checkbox to propagate initial parameter estimates across all sort levels. Users can select this option when more than one sort variable is used in the InVitro Data input dataset. If this option is selected, then initial estimates and boundaries are entered or mapped only for the first sort level. The final parameter estimates from the first sort level provide the initial estimates for each consecutive sort level.
-
Specify how to determine bounds:
•Select User Supplied Bounds to enter lower and upper bounds for each parameter estimate. Enter the boundary values in the Lower and Upper columns in the InVitro Estimates panel. Phoenix uses curve stripping to provide initial estimates. If curve stripping fails, then Phoenix uses the grid search method.
•Select Phoenix bounds to have Phoenix generate the lower and upper bounds for each parameter. Phoenix uses curve stripping to provide initial estimates, and then applies boundaries to the model parameters for model fitting. If curve stripping fails, the model fails because Phoenix cannot use grid search for initial estimates without user-supplied boundaries.
•Select Do Not Use Bounds to not apply lower or upper boundaries to parameter estimates.
Note:When Use raw data is selected, all dissolution model options are made unavailable. Also, the InVitro Estimates panel is removed from the Setup tab.
-
Click Fit Dissolution Data to run the model and generate output.
-
Click OK in the completion dialog.
Use the Main Mappings panel to identify how input variables are used in a IVIVC object. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Sort: Individual profiles used to sort the output.
•Independent: Sampling time values. Column type must be numeric.
•Values: Concentration sampling values.
•InVivo Formulation: Drug formulation identifiers.
Caution:“Formulation” is a reserved word for InVivo Data. If a column named “Formulation” is mapped to a Sort variable, it can prevent the object from performing a prediction. In such a case, the IVIVC object must be recreated, settings reapplied, the Formulation column’s name changed, remapped, and the object re-executed.
Users can specify dosing amounts and infusion times in the InVivo dosing panel. Enter the dosage amount for each formulation in the InVivo Dosing panel. For infusions, enter the infusion time for each formulation. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•InVivo Dose: Dose amount.
•InVivo Dose Formulation: The formulation associated with each dose amount.
•Duration: (Available when Infusion is selected as the Reference Data Type in the InVivo tab) Length of time for the infusion.
The InVivo tab includes settings to identify time-concentration or absorption data, formulations, and dosages. Use this tab to fit a unit impulse response (UIR) function to reference concentration data, and/or to deconvolve an existing UIR function and concentration data in order to estimate the fraction of drug absorbed over time.
•Generating UIR function and deconvolving data
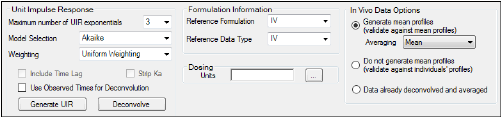
The IVIVC object can compute UIRs for all profiles based on the selected reference formulation. It fits a polyexponential function to each profile and chooses the one with the best fit based on either Akaike Information Criterion (AIC) or Weighted SSR.
-
In the Maximum number of UIR exponentials menu, select the number of exponentials to use in the UIR function.
-
In the Model Selection menu, select the whether to use AIC or Weighted SSR in the UIR function.
-
In the Weighting menu, select one of five methods for weighting data during modeling (See “Weighting” in the WinNonlin NCA section for explanations of the various schemes).
•Uniform Weighting
•1/Y (weight by 1/observed Y)
•1/Yhat (weight by 1/predicted Y [iterative reweighting])
•1/(Y*Y) (weight by 1/observed Y2)
•1(Yhat*Yhat) (weight by 1/predicted Y2 [iterative reweighting])
If oral reference formulation and data type is selected in the Formulation Information area, then the Include Time Lag and Strip Ka checkboxes are made available.
-
Check the Time Lag checkbox to include a lag time parameter in the UIR model.
-
Check the Strip Ka checkbox to generate the polyexponential describing the IV bolus pharmacokinetics. This assumes that absorption is truly first-order, fits a model that is n-compartment polyexponential, and convolved with first-order absorption. The absorption is mathematically separated from the decay. If this option is not selected, then the UIR is a polyexponential model of the reference data.
-
Check the Use Observed Times for Deconvolution checkbox to create deconvolved data points (fraction absorbed) only at the times at which observations are recorded in the In Vivo dataset. If left unselected, the deconvolution output includes the number of points specified in the Options tab, listed from initial to final observed sample times.
In this area, users can select the reference formulation and data type.
-
Reference Formulation menu: Select the formulation identifier to be used in computing the UIR and in building IVIVC models. The reference formulation must use all oral, IV bolus or IV infusion input. The IVIVC object does not support combinations of inputs for UIR calculation.
-
Reference Data Type menu: Select whether the reference input is oral, IV bolus or IV infusion.
-
In the Dosing Units field, type the dosing units to use with the PK data.
Or
Click the Dosing Units button to use the Units Builder dialog to add dosing units.
button to use the Units Builder dialog to add dosing units.
Caution:If you are using an external worksheet for the InVivo dosing data, make sure to map the worksheet before using the Dosing Units field. If you enter a unit in the field first that is different from the one in the worksheet, the unit used for the Prediction Dosing data will not get updated correctly. If this happens you will need to re-enter the information in the Prediction Dosing internal worksheet.
See “Using the Units Builder” for more details on this tool.
In this area, users can choose from three averaging options for observed, or InVivo, data.
-
Generate mean profiles (validate against mean profiles): The default option. When the InVivo dataset contains multiple subjects per formulation selecting this option creates a mean profile for each formulation. The mean profile is used for validation purposes, but is not used for model building.
•In the Averaging menu, select Mean to generate arithmetic means or GeoMean to generate geometric means.
Note:Changing the averaging method used to validate mean profiles affects the Generate UIR, Validate Correlation and Predict PK processes. It does not affect the Deconvolution or Build Correlation processes, because the mean profile is only used for validation.
-
Do not generate mean profiles (validate against individuals’ profiles): With this option, the AUClast and Cmax values over the predicted and observed individual profiles are averaged and then the %PE is calculated based on those averages.
-
Data already deconvolved and averaged: If the selected input dataset has already been deconvolved and averaged, choose this option to have Phoenix skip these steps. No UIR generation, deconvolution, or averaging is performed on the input dataset when the this option is selected. Data that is already deconvolved and averaged is validated against individual profiles.
Note:If this option is selected, then sort keys such as subject or study ID must be included in the datasets mapped to the InVivo Data and UIR panels, even if only mean data is used. If no sort column exists in those datasets, users must add a sort column. The sort column must contain the UIR parameters that are used with the corresponding plasma profile. For example, if only one UIR function is used for all formulations, then a sort column containing the parameter A1 must be added to the InVivo and UIR datasets.
Selecting Data already deconvolved and averaged causes the following four actions to occur:
•Phoenix displays a message warning users that any current deconvolution output in the IVIVC project will be removed:
•Two new panels, UIR and InVivo Fit, are made available in the Setup tab. For instructions on using the UIR and InVivo Fit panels, see “UIR panel” and “InVivo Fit panel”.
•All of the Unit Impulse Response options are made unavailable to the user.
•The Vivo.Fa_Avg and Vivo.Fa_Avg By Profile plots are not created. If they already exist in the IVIVC results, they are removed.
Note:If any IVIVC operations are dependent on deconvolved and averaged data, and the averaging option is changed, then the IVIVC process that contains the dependent operation turns yellow in the Status Panel.
Changing any of the In Vivo Data options after the Validate Correlation step is performed, results in a message to warn users that mean profile information will be removed from the project:
Generating UIR function and deconvolving data
-
When all InVivo tab settings are ready, click Generate UIR to fit the unit impulse response function(s) and generate the InVivo worksheet and plot results.
Note:If the UIR step should fail due to incorrect column mappings (for example, the Sort and Values column were inadvertently switched), the Validate Correlation and Predict PK steps will continue to fail, even after the InVivo Data mappings are corrected. Make a copy of the InVivo dataset, add a prefix to all of the column names (e.g., change the “Time” column to “XTime”), then map this dataset to the InVivo Data setup and map columns appropriately. The Validate Correlation and Predict PK steps will now execute successfully
-
When UIR generation is complete, click OK in the completion dialog.
-
Click Deconvolve to compute the fraction absorbed over time and to perform numeric deconvolution.
-
When deconvolution is complete, click OK in the completion dialog.
The IVIVC object uses the subject and formulation sort keys specified to determine individual profiles to deconvolve, using the dosing information and the UIRs just generated. It generates estimates for the fraction absorbed at 101 (default value) time points as specified in the Option tab, unless the Use Observed Times for Deconvolution checkbox is selected.
Note:If users have already determined the UIR and (average) Fraction Absorbed, then all InVivo settings must still be specified in order to generate the correct output.
-
Select the Data already deconvolved and averaged option button in the In Vivo Data Options area to skip UIR generation and deconvolution.
This panel is only available if Data already deconvolved and averaged is selected in the InVivo tab. Users should map a worksheet that contains the UIR polyexponential parameters in a stacked format to use with the IVIVC object. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Sort: The same sort columns used in the InVivo dataset. The sort columns identify unique individuals.
•InVivo Formulation: Drug formulation identifiers.
•Parameter: IVIVC model parameters for each formulation.
•Estimate: Estimated parameter values.
This panel is only available if Data already deconvolved and averaged is selected in the InVivo tab. In the InVivo Fit panel, map a dataset that contains the mean deconvolved absorption profiles for each formulation in a stacked format. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•InVivo Formulation: Drug formulation identifiers.
•Independent: Sampling time values.
•Mean: Mean deconvolved absorption data.
The Correlation tab and Correlation Estimates panel support the creation and validation of an InVitro-InVivo correlation model for the dissolution data mapped to the InVitro Data panel, and the absorption data that is either generated from the deconvolution step in the InVivo tab or is mapped to the InVivo Fit panel.
The correlation is built using the formulations identified as Internal in the InVitro Formulation tab. The IVIVC object fits the selected correlation model to absorption data using the dissolution profiles from the InVitro Data panel as inputs to the correlation function.
To validate the correlation, Phoenix can estimate AUCs and maximum concentrations based on the model predictions, and compare the estimates to actual values from the InVivo data.
To perform the estimations and comparison, Phoenix convolves predicted fractions absorbed and UIRs, then runs noncompartmental analysis on the predicted PK data and the observed InVivo data.
In the Correlation Estimates panel, enter the initial values or lower and upper boundaries for the model parameters.
See “Correlation tab” for instructions on setting correlation model parameter options.
Depending on the selections made in the Correlation tab, context associations for Correlation Estimates can include the following. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Parameter: Correlation model parameters.
•Initial: Initial estimates for the parameter.
•Lower: Lower boundary used for parameter estimates.
•Upper: Upper boundary used for parameter estimates.
The values entered in the Plot Values panel apply only to the Tvivo vs. Tvivo plot (Levy.Tvivo vs Tvitro in the results). This plot is created by selecting the Tvivo vs. Tvitro (Levy) option in the Correlation tab. The Tvivo vs. Tvitro plot finds the times for dissolution and absorption for a matching fraction of dissolution and absorption. The absorption and dissolution values used to match the time values can be entered in the Plot Values panel. The default values are zero to one, incremented by 0.1. Users can change the default values if needed. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Interpolation Values: Interpolation values used to plot InVivo and InVitro time values at specified intervals.
The Correlation tab allows users to select the correlation model to use to fit fraction absorbed (Fabs).
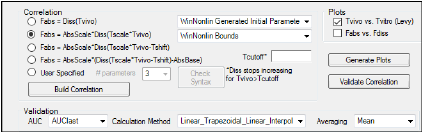
Correlation area
-
Select the correlation model to use to fit the fraction absorbed (Fabs).
•Fabs=Diss(Tvivo)
•Fabs=AbsScale*Diss(Tscale*Tvivo)
•Fabs=AbsScale*Diss(Tscale*Tvivo – Tshift)
•Fabs=AbsScale*(Diss(Tscale*Tvivo – Tshift) – AbsBase)
-
Select the User Specified option button to create a custom correlation model using the WinNonlin ASCII modeling language.
If the User Specified option button is selected, then the Correlation.Correlation.WNL5 ASCII Format panel is made available in the Setup tab. Users can write a correlation model using the legacy WinNonlin ASCII modeling language in this panel.
•If a user-specified correlation is used, click Check Syntax to verify the WNL5 ASCII code.
•In the # parameters menu, select the number of model parameters.
-
In the Tcutoff field (optional), enter the in vivo time at which the dissolution and absorption will stop increasing. That is, for the built-in models for Correlation, enter “Tvivo” at which the “Diss” function stops increasing and, therefore, the “Fabs” function also stops increasing. In other words, if Tvivo > Tcutoff, then Diss (Tvivo)=Diss(Tcutoff) and Fabs(Tvivo)=Fabs(Tcutoff).
-
Specify initial value options for correlation model parameters.
•Select User Supplied Initial Parameter Values to enter initial parameter estimates in the Correlation Estimates panel.
•Select WinNonlin Generated Initial Parameter Values to have Phoenix determine the initial parameter values.
-
Specify boundary options for correlation model parameters.
•Select User Supplied Bounds to enter lower and upper bounds for each parameter estimate. Enter the boundary values in the Lower and Upper columns in the Correlation Estimates panel.
•Select Phoenix bounds to have Phoenix generate the lower and upper bounds for each parameter.
•Select Do Not Use Bounds to not apply lower or upper boundaries to parameter estimates.
-
Click Build Correlation to run the correlation model.
-
When correlation modeling is complete, click OK in the completion dialog.
Validation area
-
Select the type of area under the curve to compute in the validation process from the AUC menu.
•AUClast: dose time through the last positive concentration value (AUClast).
•AUCall: from dose time through the final observation time (AUCall).
•AUCINF_obs: AUClast+Clast/Lambda Z, where Clast is the last observed concentration (AUCINF_obs).
•AUCINF_pred: AUClast+Clast/Lambda Z, where Clast is the last predicted concentration (AUCINF).
-
In the Calculation Method menu, select an AUC calculation method.
The selected calculation method applies to AUC computations. All methods reduce to the log trapezoidal rule, the linear trapezoidal rule, or both. See “Partial area calculation” for descriptive equations of the calculation methods. Note that the interpolation options of the AUC calculation methods have no bearing on the validation calculations because partial areas are not computed.
•Linear_Log_Trapezoidal: Uses the log trapezoidal rule after Cmax, or after C0 for IV bolus if C0 > Cmax. Otherwise the linear trapezoidal rule is used. If Cmax is not unique, then the first maximum is used. This method uses linear trapezoids before Tmax and log trapezoids after Tmax.
•Linear_Trapezoidal_Linear_Interpolation: Applies the linear trapezoidal rule to each pair of consecutive points in the dataset that have non-missing values, and sums up these areas. This method uses linear trapezoids before and after Tmax.
•Linear_Up_Log_Down: The linear trapezoidal rule is used any time that the concentration data is increasing, and the logarithmic trapezoidal rule is used any time that the concentration data is decreasing. This method uses linear trapezoids up and logarithmic trapezoids down before Tmax and linear trapezoids up and logarithmic trapezoids down after Tmax.
Note:The Linear_Log_Trapezoidal and Linear_Up_Log_Down methods apply the same exceptions in area calculation. If a Y value (concentration, rate, or effect) is less than or equal to zero, Phoenix defaults to the linear trapezoidal rule for that point. If adjacent Y values are equal to each other, Phoenix defaults to the linear trapezoidal rule.
-
In the Averaging menu, select Mean to generate arithmetic means or GeoMean to generate geometric means.
-
Click Validation Correlation to generate a table of prediction errors for AUC and Cmax:
-
When correlation validation is complete, click OK in the completion dialog.
A worksheet called Validation.Validation Errors is created in the Results tab that contains prediction errors for AUC and Cmax. The %PE column in the Validation.Validation Errors worksheet represents the prediction error percentage.
•Prediction error percentage is calculated as %PE (prediction error)=(Predicted-Observed) / Observed x 100%.
•For the Avg Internal formulation, the average is taken over the absolute values of PE for the formulations specified as Internal.
Plots area
Two diagnostic plots can be generated that can help with selecting a correlation model. Select the checkbox beside a plot to include it in the results. To see a final correlation plot rather than a preliminary diagnostic plot, view the Correlation.Abs vs Diss.Corr Overlay plot after completing the Build Correlation step.
-
Check the Tvivo vs. Tvitro (Levy) checkbox to include a Levy plot of in-vitro versus in-vivo times for a given fraction that is absorbed or dissolved (Levy Plot.Tvivo vs Tvitro).
-
Check the Fabs vs. Fdiss checkbox to include a plot of fraction absorbed vs. fraction dissolved at corresponding times, sorted by formulation (Levy Plots.Fabs vs Fdiss).
-
Click Generate Plots to create the plots.
-
When the plots are generated, click OK in the completion dialog.
The default for the regression line in the Levy Plot is to fix the intercept at the origin. To calculate the intercept, do the following:
-
From the Results tab, double-click the Levy Plot to display it as a separate window.
-
In the tree view, select Graphs > Levy Plot > Regression.
-
Unselect the Fix intercept at origin checkbox.
Select the dataset that contains the percent dissolution data, as a fraction dissolved over time, and the corresponding time points and formulation identifiers. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Time: Predicted sampling time values. Column type must be numeric.
•Dissolution: Dissolution data.
•Formulation: Formulation identifiers.
Note:Be sure to complete mapping of Prediction Data prior to mapping external worksheet data to Prediction Estimates. Otherwise, the validation fails.
In cases where a prediction has been partially set up at one time and then removed, the IVIVC object will appear as being out-of-date, even though there is no Prediction output yet.
The Prediction Dosing panel allows users to specify dosing options for the test formulation. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Prediction Dose: Dose amount used in the prediction.
•Prediction Formulation: Test formulation from the prediction data.
Caution:If you are using an external worksheet for the InVivo dosing data, make sure to map the worksheet before using the Dosing Units field. If you enter a unit in the field first that is different from the one in the worksheet, the unit used for the Prediction Dosing data will not get updated correctly. If this happens you will need to re-enter the information in the Prediction Dosing internal worksheet.
The Prediction Estimates panel allows users to specify initial estimates and lower and upper boundaries for test formulation parameters. Users can also specify whether a parameter is fixed or estimated. See “Prediction tab” for initial value, and lower and upper bound options.
Depending on the model, users can fix certain parameters to known values. For example, the fraction dissolved, Finf, can be fixed to 1.0 to prevent incomplete measurements from biasing other parameter estimates.
All iterative estimation procedures require initial estimates of the parameters. Phoenix can generate initial estimates for built-in models, or users can provide them.
For IVIVC models only, some parameters can be assigned fixed values while others are estimated. The fixed command can be used for both modeling and simulating. Fixed parameters use a set value, so are no longer considered parameters. This reduces the number of parameters in the model. Statistics such as VIF are not computed for fixed parameters.
Note:If the Prediction Estimates data source is an external worksheet, be sure to complete the mapping of Prediction Data prior to mapping Prediction Estimates. Otherwise, the validation fails.
Depending on the selections made in the Prediction tab, context associations for Prediction Estimates can include the following. Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Sort: The test formulation names that are listed in the Prediction dataset. There can be multiple test formulations used as sort values. This mapping is required. Note that formulation names must be mapped to the Sort context.
•Parameter: Test formulation parameters.
•Initial: Initial estimates for the parameter.
•Fixed or Estimated: Whether or not the initial parameter is fixed or estimated.
•Lower: Lower boundary used for parameter estimates.
•Upper: Upper boundary used for parameter estimates.
In order to provide smoothing, users can fit the dissolution data to a sigmoidal dissolution model. The Prediction tab generates a table of prediction errors for AUC and Cmax by comparing predicted data for test formulations mapped to the Prediction Data panel to the target formulation identified on the InVitro Formulation panel.
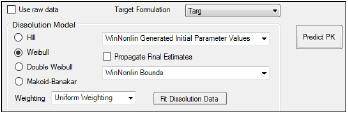
Prediction tab
The options available in this tab are the same as those in the InVitro tab (see “InVitro tab” for descriptions), with the exception of the following:
•Target Formulation: Select a target formulation to use as the comparator in predictions. Only one formulation can be selected in the menu.
•Predict PK: Generate the prediction error worksheets. Worksheets are created for AUC, Cmax and other NCA parameters. This is done by comparing test formulations to the target formulation identified on the InVivo Formulation panel. Typical NCA output like final parameters and summary table worksheets are created in the Results tab.
Note:Even if a prediction is not being done, the Target Formulation should be set in order to use output from IVIVC downstream. If it is not set, it will prevent IVIVC from passing verification, which will prevent use of output in other objects.
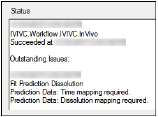
The Status tab lists whether or not modeling results were successful, and any errors in model option specifications.
The Options tab allows users to specify model runtime options and the number of output points.
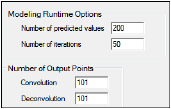
Options tab
-
In the Number of predicted values field, type the number of predicted values that are generated by IVIVC modeling operations.
-
In the Number of iterations field, type the maximum number of iterations to use during model fitting.
-
In the Convolution Output Points field, type the number of output points to use in the convolution output.
-
In the Deconvolution Output Points field, type the number of output points to use in the deconvolution output.
Users can typically leave these options set to their default values.
