Results
Select the Results tab to view the output. IVIVC object output includes worksheets, plots, and text files. The lists of output in this section are grouped into tables based on the process by which they are generated.
•Build correlation process output
•Deconvolve data process output
•Deconvolve process output
•Output for the fit dissolution step of Prediction
•Output for InVitro stage of analysis
•Output for Levy plot results of Percent absorbed vs percent dissolved
•UIR Modeling output for InVivo stage
•Predict PK process output
•Validate correlation process output
Settings are not recorded on the History tab upon execution. However, they can be found in the individual Settings text output files for the different steps.
Note:Word Export is not recommended for IVIVC as the export would include all internal/hidden steps whose results are not readily discernible to the user.
Build correlation process output
|
Worksheet Name
|
Description
|
|
Correlation.Abs vs Diss data
|
Fraction Absorbed vs Dissolved Data
|
|
Correlation.Correlation Overlay data
|
Fraction Absorbed vs Dissolved Overlay Data
|
|
Correlation.Correlation.Condition Numbers
|
Correlation Modeling Results
|
|
Correlation.Correlation.Correlation Matrix
|
|
Correlation.Correlation.Diagnostics
|
|
Correlation.Correlation.Differential Equations
|
|
Correlation.Correlation.Dosing Used
|
|
Correlation.Correlation.Eigenvalues
|
|
Correlation.Correlation.Final Parameters
|
|
Correlation.Correlation.Final Parameters Pivoted
|
|
Correlation.Correlation.Initial Estimates
|
|
Correlation.Correlation.Minimization Process
|
|
Correlation.Correlation.Partial Derivatives
|
|
Correlation.Correlation.Predicted Data
|
|
Correlation.Correlation.Secondary Parameters
|
|
Correlation.Correlation.Secondary Parameters Pivoted
|
|
Correlation.Correlation.Stacked Partial Derivatives
|
|
Correlation.Correlation.Summary Table
|
|
Correlation.Correlation.User Defined Settings
|
|
Correlation.Correlation.VarianceCovariance Matrix
|
|
Correlation.Model Sim Vitro
|
Correlation Modeling Simulated Vitro Input
|
|
Correlation.Model Sim Vivo
|
Correlation Modeling Simulated Vivo Input
|
|
Correlation.Vitro Vivo Times
|
Used for in generating the Fraction Absorbed vs Dissolved plot.
The tVitro added to Fa-Avg is based on the correlation function, all columns are removed except for formulation, tVivo, tVitro, and Mean
|
|
Plot Name
|
Description
|
|
Correlation.Abs vs Diss Corr Overlay
|
Fraction Absorbed vs Dissolved with Correlation Overlay data.
This plot is generated for built-in models, but not for user models.
|
|
Correlation.Correlation.Observed Y and Predicted Y vs X
|
Correlation Modeling Results
|
|
Correlation.Correlation.Partial Derivatives Plot
|
|
Correlation.Correlation.Predicted Y vs X
|
|
Correlation.Correlation.Weighted Residual Y vs X
|
|
Text File Name
|
Description
|
|
Correlation.Correlation.Core output
|
Correlation Modeling Results
|
|
Correlation.Correlation.Settings
|
Deconvolve data process output
|
Worksheet Name
|
Description
|
|
Deconvolution Input
|
Input to the deconvolution step
|
Deconvolve process output
|
Worksheet Name
|
Description
|
|
Vivo.Deconvolution.Dosing
|
Deconvolution modeling output for InVivo stage of analysis
|
|
Vivo.Deconvolution.Exponential Terms
|
|
Vivo.Deconvolution.Fa-avg
|
Fa-Avg plot data
|
|
Vivo.Deconvolution.Fitted Values
|
Deconvolution modeling output for InVivo stage of analysis
|
|
Vivo.Deconvolution.Parameters
|
|
Vivo.Deconvolution.Values
|
|
Plot Name
|
Description
|
|
Vivo.Deconvolution.Fa-Av
The plot is not created if the Data already deconvolved and averaged option is selected, or if no sort variables are selected in the In-Vivo tab.
|
Fa-Avg overlaid.
Average fraction absorbed by formulation.
For the chart, Fabs grouped by subject with Fabs_avg overlaid and sorted by formulation
|
|
Vivo.Deconvolution.Fa-Av By Profile
The plot is not created if the Data already deconvolved and averaged option is selected, or if no sort variables are selected in the In-Vivo tab.
|
Fa-Avg plot by profile
Fabs grouped by formulation, vs. time, and sorted by profile
|
|
Vivo.Deconvolution.Fitted Curves Plot
|
Deconvolution modeling output for InVivo stage of analysis
|
|
Vivo.Deconvolution.Input Rates Plot
|
|
Text File Name
|
Description
|
|
Vivo.Deconvolution.Fa-avg
|
Descriptive Stats settings for Fa-Avg
|
|
Vivo.Deconvolution.Settings
|
Deconvolution modeling output for InVivo stage of analysis
|
Output for the fit dissolution step of Prediction
|
Worksheet Name
|
Plot Name
|
|
Prediction.Diss.Condition Numbers
|
Prediction.Diss.Observed Y and Predicted Y vs X
|
|
Prediction.Diss.Correlation Matrix
|
Prediction.Diss.Partial Derivatives Plot
|
|
Prediction.Diss.Diagnostics
|
Prediction.Diss.Predicted Y vs Observed Y
|
|
Prediction.Diss.Differential Equations
|
Prediction.Diss.Predicted Y vs X
|
|
Prediction.Diss.Eigenvalues
|
Prediction.Diss.Residual Y vs Predicted Y
|
|
Prediction.Diss.Final Parameters
|
Prediction.Diss.Residual Y vs X
|
|
Prediction.Diss.Final Parameters Pivoted
|
Text File Name
|
|
Prediction.Diss.Initial Estimates
|
Prediction.Diss.Core output
|
|
Prediction.Diss.Minimization Process
|
Prediction.Diss.Settings
|
|
Prediction.Diss.Partial Derivatives
|
|
|
Prediction.Diss.Predicted Data
|
|
Prediction.Diss.Secondary Parameters
|
|
Prediction.Diss.Secondary Parameters Pivoted
|
|
Prediction.Diss.Stacked Partial Derivatives
|
|
Prediction.Diss.Summary Table
|
|
Prediction.Diss.User Defined Settings
|
|
Prediction.Diss.VarianceCovariance Matrix
|
Output for InVitro stage of analysis
|
Worksheet Name
|
Plot Name
|
|
Vitro.Dissolution.Condition Numbers
|
Vitro.Dissolution.Observed Y and Predicted Y vs X
|
|
Vitro.Dissolution.Correlation Matrix
|
Vitro.Dissolution.Partial Derivatives Plot
|
|
Vitro.Dissolution.Diagnostics
|
Vitro.Dissolution.Predicted Y vs Observed Y
|
|
Vitro.Dissolution.Differential Equations
|
Vitro.Dissolution.Predicted Y vs X
|
|
Vitro.Dissolution.Eigenvalues
|
Vitro.Dissolution.Residual Y vs Predicted Y
|
|
Vitro.Dissolution.Final Parameters
|
Vitro.Dissolution.Residual Y vs X
|
|
Vitro.Dissolution.Final Parameters Pivoted
|
Vivo.Deconvolution.Cumulative Rates Plot
|
|
Vitro.Dissolution.Initial Estimates
|
Text File Name
|
|
Vitro.Dissolution.Minimization Process
|
Vitro.Dissolution.Core output
|
|
Vitro.Dissolution.Partial Derivatives
|
Vitro.Dissolution.Settings
|
|
Vitro.Dissolution.Predicted Data
|
|
|
Vitro.Dissolution.Secondary Parameters
|
|
Vitro.Dissolution.Secondary Parameters Pivoted
|
|
Vitro.Dissolution.Stacked Partial Derivatives
|
|
Vitro.Dissolution.Summary Table
|
|
Vitro.Dissolution.User Defined Settings
|
|
Vitro.Dissolution.VarianceCovariance Matrix
|
Output for Levy plot results of Percent absorbed vs percent dissolved
|
Worksheet Name
|
Plot Name
|
|
Levy Plots.%Abs vs %Diss.Levy Plot Values
|
Levy Plots.%Abs vs %Diss
|
|
Levy Plots.%Abs vs %Diss.Parameters
|
Levy Plots.Fabs vs Fdiss
|
|
Levy Plots.Fabs vs Fdiss.Levy Plot Values
|
|
|
Levy Plots.Fabs vs Fdiss.Parameters
|
UIR Modeling output for InVivo stage
|
Worksheet Name
|
Plot Name
|
|
Vivo.UIR.Condition Numbers
|
Vivo.UIR.Observed Y and Predicted Y vs X
|
|
Vivo.UIR.Correlation Matrix
|
Vivo.UIR.Partial Derivatives Plot
|
|
Vivo.UIR.Diagnostics
|
Vivo.UIR.Predicted Y vs Observed Y
|
|
Vivo.UIR.Differential Equations
|
Vivo.UIR.Predicted Y vs X
|
|
Vivo.UIR.Dosing Used
|
Vivo.UIR.Residual Y vs Predicted Y
|
|
Vivo.UIR.Eigenvalues
|
Vivo.UIR.Residual Y vs X
|
|
Vivo.UIR.Final Parameters
|
Test File Name
|
|
Vivo.UIR.Final Parameters Pivoted
|
Vivo.UIR.Core output
|
|
Vivo.UIR.Initial Estimates
|
Vivo.UIR.Settings
|
|
Vivo.UIR.Minimization Process
|
Vivo.UIR.Warnings and Errors
|
|
Vivo.UIR.Partial Derivatives
|
|
|
Vivo.UIR.Predicted Data
|
|
Vivo.UIR.Secondary Parameters
|
|
Vivo.UIR.Secondary Parameters Pivoted
|
|
Vivo.UIR.Stacked Partial Derivatives
|
|
Vivo.UIR.Summary Table
|
|
Vivo.UIR.User Defined Settings
|
|
Vivo.UIR.VarianceCovariance Matrix
|
Predict PK process output
|
Worksheet Name
|
Description
|
|
Prediction.Baseline Stats
|
Observed summary stats used to calculate the errors
|
|
Prediction.Conv.Results
|
Simulation Convolution modeling output for Prediction Stage
|
|
Prediction.Corr Sim.Condition Numbers
|
Correlation modeling simulation output for Prediction step
|
|
Prediction.Corr Sim.Correlation Matrix
|
|
Prediction.Corr Sim.Diagnostics
|
|
Prediction.Corr Sim.Differential Equations
|
|
Prediction.Corr Sim.Dosing Used
|
|
Prediction.Corr Sim.Eigenvalues
|
|
Prediction.Corr Sim.Final Parameters
|
|
Prediction.Corr Sim.Final Parameters Pivoted
|
|
Prediction.Corr Sim.Initial Estimates
|
|
Prediction.Corr Sim.Minimization Process
|
|
Prediction.Corr Sim.Partial Derivatives
|
|
Prediction.Corr Sim.Predicted Data
|
|
Prediction.Corr Sim.Secondary Parameters
|
|
Prediction.Corr Sim.Secondary Parameters Pivoted
|
|
Prediction.Corr Sim.Stacked Partial Derivatives
|
|
Prediction.Corr Sim.Summary Table
|
|
Prediction.Corr Sim.User Defined Settings
|
|
Prediction.Corr Sim.VarianceCovariance Matrix
|
|
Prediction.Model Sim Vitro
|
Vitro data used for Correlation Simulation
|
|
Prediction.Model Sim Vivo
|
Vivo data used for Correlation Simulation
|
|
Prediction.Observed Baseline Nca.Dosing Used
|
Observed NCA results
|
|
Prediction.Observed Baseline Nca.Exclusions
|
|
Prediction.Observed Baseline Nca.Final Parameters
|
|
Prediction.Observed Baseline Nca.Final Parameters Pivoted
|
|
Prediction.Observed Baseline Nca.Partial Area Labels
|
|
Prediction.Observed Baseline Nca.Plot Titles
|
|
Prediction.Observed Baseline Nca.Slopes Settings
|
|
Prediction.Observed Baseline Nca.Summary Table
|
|
Prediction.Predicted Nca prediction.Dosing Used
|
Predicted NCA results
|
|
Prediction.Predicted Nca prediction.Exclusions
|
|
Prediction.Predicted Nca prediction.Final Parameters
|
|
Prediction.Predicted Nca prediction.Final Parameters Pivoted
|
|
Prediction.Predicted Nca prediction.Partial Area Labels
|
|
Prediction.Predicted Nca prediction.Plot Titles
|
|
Prediction.Predicted Nca prediction.Slopes Settings
|
|
Prediction.Predicted Nca prediction.Summary Table
|
|
Prediction.Predicted Stats
|
Predicted summary stats used to calculate the errors
|
|
Prediction.Prediction Errors
|
Prediction Errors
|
Validate correlation process output
|
Worksheet Name
|
Description
|
|
Validation.Average Vivo.Statistics
|
Validation summary stats used to calculate the errors
|
|
Validation.Avg PE Stats
|
Validation percent error calculations
|
|
Validation.Baseline Stats
|
Observed summary stats used to calculate Validation errors
|
|
Validation.Conv.Exponential Terms
|
Simulation Convolution modeling output for Validation Stage
|
|
Validation.Conv.Results
|
|
Validation.Corr Sim.Condition Numbers
|
Simulation Correlation modeling output for Validation Stage
|
|
Validation.Corr Sim.Correlation Matrix
|
|
Validation.Corr Sim.Diagnostics
|
|
Validation.Corr Sim.Differential Equations
|
|
Validation.Corr Sim.Dosing Used
|
|
Validation.Corr Sim.Eigenvalues
|
|
Validation.Corr Sim.Final Parameters
|
|
Validation.Corr Sim.Final Parameters Pivoted
|
|
Validation.Corr Sim.Initial Estimates
|
|
Validation.Corr Sim.Minimization Process
|
|
Validation.Corr Sim.Partial Derivatives
|
|
Validation.Corr Sim.Predicted Data
|
|
Validation.Corr Sim.Secondary Parameters
|
|
Validation.Corr Sim.Secondary Parameters Pivoted
|
|
Validation.Corr Sim.Stacked Partial Derivatives
|
|
Validation.Corr Sim.Summary Table
|
|
Validation.Corr Sim.User Defined Settings
|
|
Validation.Corr Sim.VarianceCovariance Matrix
|
|
Validation.Model Sim Vitro
|
Correlation model simulation Vitro input for Validation Stage
|
|
Validation.Model Sim Vivo
|
|
Validation.Observed Baseline Nca.Dosing Used
|
Observed NCA analysis for Validation Stage
|
|
Validation.Observed Baseline Nca.Exclusions
|
|
Validation.Observed Baseline Nca.Final Parameters
|
|
Validation.Observed Baseline Nca.Final Parameters Pivoted
|
|
Validation.Observed Baseline Nca.Partial Area Labels
|
|
Validation.Observed Baseline Nca.Plot Titles
|
|
Validation.Observed Baseline Nca.Slopes Settings
|
|
Validation.Observed Baseline Nca.Summary Table
|
|
Validation.Predicted PK Nca.Dosing Used
|
Predicted NCA analysis for Validation Stage
|
|
Validation.Predicted PK Nca.Exclusions
|
|
Validation.Predicted PK Nca.Final Parameters
|
|
Validation.Predicted PK Nca.Final Parameters Pivoted
|
|
Validation.Predicted PK Nca.Partial Area Labels
|
|
Validation.Predicted PK Nca.Plot Titles
|
|
Validation.Predicted PK Nca.Slopes Settings
|
|
Validation.Predicted PK Nca.Summary Table
|
|
Validation.Predicted Stats
|
Predicted summary stats used to calculate Validation errors
|
|
Validation.Validation Errors
|
Validation Errors
|
|
Plot Name
|
Description
|
|
Validation.Corr Sim.Observed Y and Predicted Y vs X
|
Simulation Correlation modeling output for Validation Stage
|
|
Validation.Corr Sim.Partial Derivatives Plot
|
|
Validation.Corr Sim.Predicted Y vs X
|
|
Validation.Corr Sim.Weighted Residual Y vs X
|
|
Validation.Observed Baseline Nca.Observed Y and Predicted Y vs X
|
Observed NCA analysis for Validation Stage
|
|
Validation.Predicted PK Nca.Observed Y and Predicted Y vs X
|
Predicted NCA analysis for Validation Stage
|
|
Validation.Val Conv Out
|
Convolution results for simulation part of Validation
|
|
Text File Name
|
Description
|
|
Validation.Baseline Stats
|
Observed summary stats settings used to calculate Validation errors
|
|
Validation.Conv.Settings
|
Simulation Convolution modeling output for Validation Stage
|
|
Validation.Corr Sim.Core output
|
Simulation Correlation modeling output for Validation Stage
|
|
Validation.Corr Sim.Settings
|
|
Validation.IVIVC.Validation
|
Settings for the filter prior to generation of Validation Errors
|
|
Validation.Observed Baseline Nca.Core output
|
Observed NCA analysis for Validation Stage
|
|
Validation.Observed Baseline Nca.Settings
|
|
Validation.Predicted PK Nca.Core output
|
Predicted NCA analysis for Validation Stage
|
|
Validation.Predicted PK Nca.Settings
|
|
Validation.Settings
|
Validation settings
|
Sigmoidal and Dissolution models
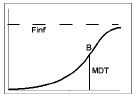
Model 601 — Hill
Example graph and equation for model 601 — Hill
where the estimated parameters are:
Finf: amount released at time infinity, using the preferred units for y
MDT: mean dissolution time, in the preferred units for x (time)
b: slope factor (no units)
int: y-intersect; zero (default) or first positive y value in data if estimated, with bounds of zero and 10 times that value
This model includes an optional lag time in the form of a step function, i.e.:
|
y(t)=(Finf *U(t – tlag)b)/(MDTb+U(t – tlag)b)
|
(1)
|
where U(x)=x if x >= 0; U(x)=0 if x < 0.
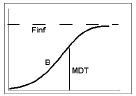
Model 602 — Weibull
Example graph and equation for model 602 — Weibull
where the estimated parameters are:
Finf: amount released at time infinity, using the preferred units for y
MDT: mean dissolution time, in the preferred units of x (time)
b: slope factor (no units)
int: y-intersect; zero (default) or first positive y value in data if estimated, with bounds of zero and 10 times that value
This model includes an optional lag time in the form of a step function, i.e.:
|
y(t)=Finf (1 – exp[–(U(t – tlag)/MDT)b])
|
(2)
|
where U(x)=x if x >= 0; U(x)=0 if x < 0.
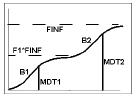
Model 603 — Double Weibull
Example graph and equation for model 603 — Double Weibull
where the estimated parameters are:
f1: weighting factor, setting the fraction due to each Weibull (no units)
Finf: amount released at time infinity, using the preferred units for y
MDT1, MDT2: mean dissolution time for each Weibull, (x units)
b1, b2: slope factors (no units)
int: y-intersect; zero (default) or first positive y value in data if estimated, with bounds of zero and 10 times that value
This model includes an optional lag time in the form of a step function, i.e.:
|
y(t)=f1 * Finf (1 – exp[–(U(t – tlag)/MDT1)b1])
+(1 – f1) * Finf (1 – exp[–(U(t – tlag)/MDT2)b2])
|
(3)
|
where U(x)=x if x >= 0; U(x)=0 if x < 0.
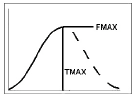
Model 604 — Makoid-Banakar
Example graph and equation for model 604 — Makoid-Banakar
where the estimated parameters are:
Fmax: maximum y value, using the preferred units for y
Tmax: time of maximum y value, using the preferred units for x (time)
b: slope factor (no units)
int: y-intersect; zero (default) or first positive y value in data if estimated, with bounds of zero and 10 times that value
This model includes an optional lag time in the form of a step function, i.e.:
|
y(t)=Fmax * [U(t – tlag)/Tmax]b * exp[b * (1 – U(t – tlag)/Tmax)]
|
(4)
|
where U(x)=x if x >= 0; U(x)=0 if x < 0.
Note:The FMAX parameter can be fixed at a certain value by doing one of the following:
– Set the initial estimates option to User-Supplied Initial Parameter Values, and then set initial estimates for any estimated parameters.
– Create a worksheet and use it as an external worksheet for the initial estimates.
ASCII user model for correlations
A custom model for an IVIVC correlation needs to include only the parameter definitions and model equations. No other statements are required.
-
In the # parameters menu in the Correlation tab, select the number of model parameters.
-
In the Correlation Estimates panel, enter an initial value for each model parameter.
-
Edit the statements in the Correlation Code panel to reflect the model. Use the ASCII model syntax and the following keywords that are specific to IVIVC:
•IVINTERP(): This function is used to interpolate dissolution data. A common usage is for X to be the in vivo time, and X is then scaled or shifted by model parameters to yield an in vitro time, which is called T. IVINTERP is then used to interpolate the dissolution data to find the dissolution at time T. The first and last values in the dissolution data are used when T is respectively before or after the time range of the dissolution data.
•DOSEVIVO: This variable contains the dose amount for each formulation. The wizard will translate this variable into the DTA array that is part of the WNL modeling language.
Data deficiencies resulting in missing values for PK parameters
Phoenix reports missing values for PK parameters in the following cases.
Cases when missing PK parameter values are reported
|
Profile issue(s)
|
Parameters reported as missing
|
|
Profile with no non-missing observations after dose time.
|
All final parameters
|
|
Bolus dosing and <=1 non-missing observation after dose time.
|
All final parameters
|
|
Blood/plasma data with no non-zero observation values.
|
All final parameters except:
Cmax, AUClast, AUCall, AUMClast
|
|
Blood/plasma data with all zero values except one non-zero value at dosing time
|
All final parameters
|