Note:To run a model through use of the nmfe script supplied with NONMEM, you have to instruct Pirana as to the location of the NONMEM installation. This is done by setting up profiles using the Settings dialog (see the “NONMEM” section of the Settings dialog description).
If you are running NONMEM through PsN, the location of NONMEM is already specified in the psn.conf file of PsN.
-
Select the model to run.
-
Right-click the selected model and choose nmfe.
Or
Click in the toolbar.
in the toolbar.
Or
Use the Control-R keyboard shortcut. -
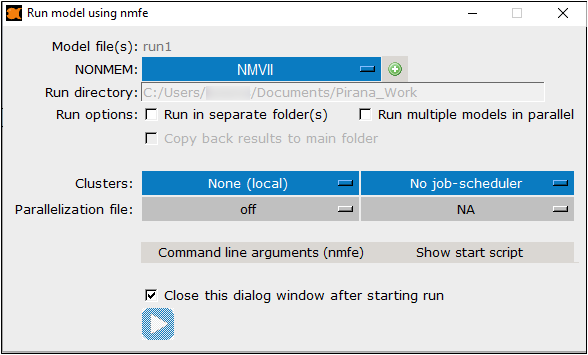
Select the NONMEM installation from the pull-down.
Click + to locate a NONMEM installation that is not listed on the pull-down. -
The Run directory shows the path specified in the Folder field on Pirana’s main page.
-
Check the Run in separate folder(s) box to create individual folders, prefixed by “nmfe_”, for each model execution.
-
Check the Run multiple models in parallel box to allow parallelized executions.
-
Check the Copy back results to main folder box to bring the results from parallel runs back to the main folder.
This option is only available when the Run multiple models in parallel box is checked. -
From the Clusters pull-down, choose None to run the job locally or choose a cluster name to run the job remotely. (Refer to “Pirana and Clusters” for details on setting up a cluster in Pirana.)
-
If the chosen cluster has job schedulers associated with it, select the scheduler to use from the second pull-down.
-
Choose the configuration file to use for the parallelization.
Parallelization configuration files can be generated using the NM parallelization file wizard in Pirana.
You can also have Pirana generate the parafile on-the-fly (select auto-MPI or auto-FPI from the Parallelization file menu). In the Settings dialog, in the NONMEM > Clusters > ‘cluster_name’ tab, set the FPI and MPI files that Pirana generates using the respective sub-tabs. (See MPI scripts settings for parallelization and FPI scripts settings for parallelization.)
Parallelization files can be imported from local or remote locations. Local import can be performed through the NONMEM option in the Software integration tab of the Settings dialog. Remote parallelization files can be imported from a cluster location defined in the NONMEM > Remote Profiles sub-tab of the Settings dialog. -
Press Command line arguments (nmfe) to toggle expanding the dialog to view and optionally add nmfe command line arguments to include in the job execution.
-
Press Show start script to toggle expanding the dialog to view and optionally edit the model execution script and command line changes for the run options selected.
-
Uncheck the Close this dialog window after starting run box to keep the dialog open.
-
Click
 to submit the job for execution.
to submit the job for execution.
The Run window contains a number of additional options for running the model.