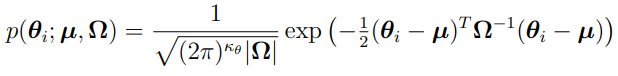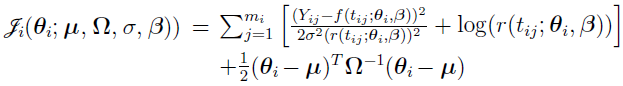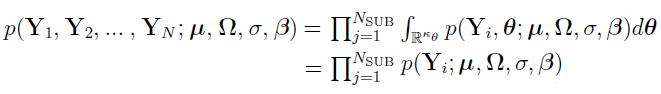Review of Laplacian-Approximation Formulation
For nonlinear mixed-effect models, there are usually two levels of variability:
intra-individual variability due to residual/measurement errors; and
inter-individual variability in parameter values due to “unexplained” variation (e.g., natural, biological variation) and/or covariates.
For example, an observable variable may be governed by the statistical model:
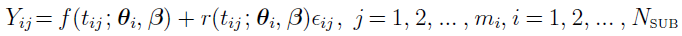
where
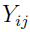 is the jth observation (e.g., drug concentration) for subject i at time
is the jth observation (e.g., drug concentration) for subject i at time 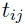 .
.
f is the function relating time 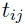 , parameter
, parameter 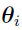 specific to subject i, and parameter
specific to subject i, and parameter 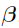 common to all subjects to observation
common to all subjects to observation 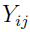 .
.
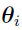 is a random vector to account for the inter-individual variability
is a random vector to account for the inter-individual variability
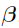 is often referred to as bare fixed effects as they are not paired with any random effects
is often referred to as bare fixed effects as they are not paired with any random effects
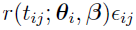 is the residual error at
is the residual error at 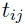 for subject i, where the functional form of r determines the type of residual error (e.g., if r ≡ 1, then it is an additive error; and if r ≡ f, then it is a proportional/multiplicative error) and random variables (
for subject i, where the functional form of r determines the type of residual error (e.g., if r ≡ 1, then it is an additive error; and if r ≡ f, then it is a proportional/multiplicative error) and random variables (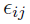 ) are assumed to be independent and identically (i.i.d.) normally distributed with zero mean and constant variance
) are assumed to be independent and identically (i.i.d.) normally distributed with zero mean and constant variance 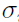 2.
2.
mi is the number of observations for subject i.
NSUB is the number of subjects.
Most PKPD population analysis are based on parametric assumptions, where the distribution of 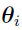 is assumed to be i.i.d. multivariate normally distributed. (Note that the distribution can also be the transformations of multivariate normal distribution.) Specifically, it is assumed that
is assumed to be i.i.d. multivariate normally distributed. (Note that the distribution can also be the transformations of multivariate normal distribution.) Specifically, it is assumed that 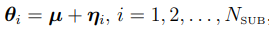 , where
, where 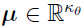 and
and 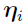 is independent and identically multi-normally distributed with zero mean and covariance matrix
is independent and identically multi-normally distributed with zero mean and covariance matrix 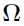 ; that is,
; that is, 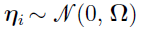 with
with 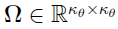 .
.
It is also assumed that 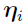 and
and 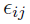 are independent. The goal of parametric methods is to find the estimates for population parameters,
are independent. The goal of parametric methods is to find the estimates for population parameters, 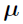 ,
, 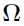 ,
, 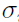 , and
, and 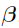 with given data. Most derivations in this section are based on work reported in reference [1].
with given data. Most derivations in this section are based on work reported in reference [1].
Let 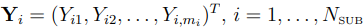 . Then, due to the i.i.d. normal assumption on
. Then, due to the i.i.d. normal assumption on 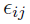 , the conditional PDF of
, the conditional PDF of 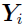 given
given 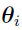 is given by
is given by
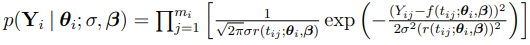
Since 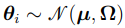 , the PDF of
, the PDF of 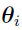 is given by
is given by
Thus the logarithm of the joint PDF of 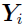 and
and 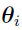 is given by
is given by
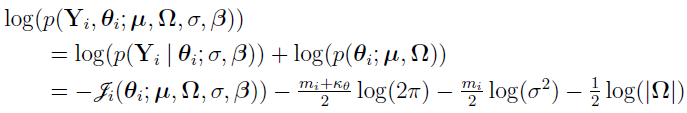
With given, 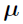 ,
, 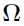 ,
, 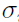 , and
, and 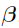 , one can optimize
, one can optimize 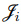 to obtain optimal individual parameter estimates for the ith individual, i = 1, 2,..., NSUB. These estimates are also referred to as conditional modes, modes of a posterior (MAP), empirical Bayes estimates (EBE) or posthoc. This is the method used to calculate individual parameter estimates for all the population analysis methods except for the EM method, which uses the conditional means rather than modes.
to obtain optimal individual parameter estimates for the ith individual, i = 1, 2,..., NSUB. These estimates are also referred to as conditional modes, modes of a posterior (MAP), empirical Bayes estimates (EBE) or posthoc. This is the method used to calculate individual parameter estimates for all the population analysis methods except for the EM method, which uses the conditional means rather than modes.
The marginal PDF of 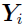 is given by
is given by 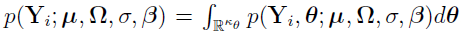 . Note that
. Note that 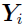 is independent (due to the independence of
is independent (due to the independence of 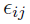 ). Hence, the marginal PDF of (
). Hence, the marginal PDF of (.png) ) is given by
) is given by
which implies that 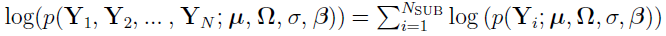 .
.
The goal of maximum likelihood methods is to maximize overall log-likelihood function (as shown in the previous equation) to obtain estimates for population parameters. Unfortunately, the marginal PDF often cannot be computed analytically due to the complexity of the involved integral. There are two main methods to solve this problem.
Approximate the required integral (e.g., through linearization, Laplacian approximation, or Gaussian quadrature method) and then maximize the resulting approximate marginal PDF to obtain population parameter estimates. The unconstrained Broyden–Fletcher–Goldfarb–Shanno (BFGS) quasi-Newton method is then often used for the corresponding optimization problem. This is the optimization algorithm used by Certara’s NLME engine.
Use the EM method, which alternates between an expectation step and a maximization step to obtain the parameter estimates that potentially will iteratively converge to the maximum likelihood estimates (refer to the “Expectation Maximization-Based Algorithms” for more information).
It is worth noting that, since 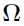 is a symmetric matrix, instead of estimating
is a symmetric matrix, instead of estimating 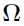 directly, one often estimates its corresponding lower triangular matrix
directly, one often estimates its corresponding lower triangular matrix 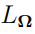 (that is,
(that is, 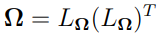 with
with 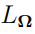 obtained by using Cholesky decomposition). This is the method used in Certara’s NLME engine.
obtained by using Cholesky decomposition). This is the method used in Certara’s NLME engine.
Before further exploration of the Laplacian-approximation-based algorithms, some additional notations should be defined. Let 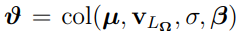 , where col is an operator that transforms a sequence of column vectors into a single column vector by stacking them one under the other. Then
, where col is an operator that transforms a sequence of column vectors into a single column vector by stacking them one under the other. Then 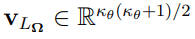 is obtained by eliminating the above diagonal elements of
is obtained by eliminating the above diagonal elements of 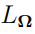 .
.
.