Commands, Arrays, and Functions
BASELINE command: Sets the (optional) baseline value for use analyzing effect data with NCA model 220.
BEGIN command: Indicates that all commands have been specified and processing should start.
BTIME command: Selects points within the Lambda Z time range to be excluded from computations of Lambda Z. In the following example, Lambda Z is to be computed using times in the range of 12 to 24, excluding the concentrations at 16 and 20 hours. These “excluded” points will, however, still be used in the computation of the pharmacokinetic parameters.
BTIME 12, 24, 2, 16, 20
If there are no times to be excluded, set numexcl to zero or leave it blank.
Do not mark a point for exclusion by setting the case weight to zero as that will also exclude this data point from the calculation of the PK parameters.
CARRYALONG command: Specifies variables from the input data grid to be copied into each of the output worksheets. These variables are not required for the analysis.
CARRY 3
CARR 1
CARRYALONG 2
See also the NCARRY command.
How Carry Along Data is Migrated to Output Multigrids
The data for carry along columns is moved to all output worksheets from PK (PK/PD) or NCA analyses, with the exception of the last worksheet, named Settings.
For all output worksheets other than the Summary Table, the carry along data will appear in the right-most column(s) of the worksheet. The first cell of data for any given profile within a carry along column will be copied over the entire profile range. There is one exception: generation of the “Summary Table” worksheets. The carry along columns will be the first columns, preceded only by any sort key columns.
Also, as there are almost the same number of observations in the raw dataset as in the Summary Table, all the cells within a profile of a carry along column are copied over to the Summary Table—not just the first value. There is one special case here as well: an NCA without a first data point of t=0, c=0. When such a data point does not exist on the raw dataset, depending on which NCA model is selected, the core program may generate a pseudo-point of t=0, c=0 to support all necessary calculations. In this event, the first cell within a carry along column is copied 'up' to the core-generated data value of 0,0.
COMMANDS (model block): The COMMANDS block allows Phoenix commands to be permanently inserted into a model. Following the COMMANDS statement, any Phoenix command such as NPARAMETERS, NCONSTANTS, PNAME, etc., may be given. To end the COMMAND section use the END statement.
COMMANDS
NCON 2
NPARM 4
PNAME 'A', 'B', 'Alpha', 'Beta'
END
CON (array): Contains the values of the constants, such as dosing information, used to define the model. The index N indicates the Nth constant.
CON(3)
The values specified by the CONSTANT command, described below, correspond to CON(1), CON(2), etc. respectively.
CONSTANTS command: Specifies the values of the constants (such as dosing information) required by the model. The arguments C1, C2, …, CN are the values.
CONSTANTS 20, 2.06, -1
CONS are 5, 2
The NCONSTANTS command must appear before CONSTANTS.
CONVERGENCE command: Specifies the convergence criterion, C. 0 < C < 0.1.
The default value is 10–4.
CONVERGENCE=1.e-6
CONV .001
The test for convergence is:
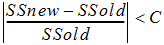
where SS is the residual sum of squares. When CONVERGENCE=0, convergence checks and halvings are turned off (useful for maximum likelihood estimates).
DATA command: Begins data input and optionally specifies an external data file. If arguments are omitted, the data are expected to follow the DATA statement. If the arguments are used, path is the full path of the dataset, up to 64 characters.
DATA
DATA 'C:\MYLIB\MYFILE.DAT'
DATE command: Places the current date in the upper right corner of each page in Core Output.
DATE
DHEADERS command: Statement indicating that data included within the model file contain variable names as the first row.
DHEADERS
If a DNAMES command is also included, it will override the names in the data.
DIFFERENTIAL model block: Statements in the DIFFERENTIAL-END block define the system of differential equations to fit. The values of the equations are assigned to the DZ array.
DIFFERENTIAL
DZ(1)=-K1+KE)*Z(1)+K2*Z(2)
DZ(2)=K1*Z(1)-K2*Z(2)
END
NDERIVATIVES must be used to set the number of differential equations to fit. NFUNCTIONS specifies the number of differential equations and/or other functions with associated data.
DINCREMENT command: Specifies the increment to use in the estimation of the partial derivatives. The argument D is the increment. 0 < D < 0.1. The default value is 0.0001
DINCREMENT is 0.001
DINC 0.005
DNAMES command: Assigns names to the columns on the dataset. The arguments 'name1', 'name2',…,'nameN' are comma- or space-separated variable names. Each may use up to eight letters, numbers or underscores, and must begin with a letter.
DNAMES 'Subject', 'Time', 'Conc'
The names appear in the output and may also be used in the modeling code. If both DNAMES and DHEADERS are included, DNAMES takes precedence.
DTA array: Returns the value of the Nth column for the current observation.
T=DTA(1)
W=DTA(3)
DTIME command: Specifies the time of the last administered dose. This value will appear on the Final Parameters table as Dosing_time. Computation of the final parameters will be adjusted for this dosing time.
DTIME 48
DZ array: Contains the current values of the differential equations. The index N specifies the equation, in the order listed in the DIFFERENTIAL block.
DZ(3)=-P(3)*Z(3)
The maximum value of N is specified by the NDERIVATIVES command.
F variable: Returns the value of the function for the current observation.
F=A*exp(-B*X)
FINISH command: Included for compatibility with PCNonlin Version 4. FINISH should immediately follow the BEGIN command.
BEGIN
FINISH
FNUMBER command: Indicates the column in the dataset that identifies data for the FUNCTION block when NFUNCTIONS > 1. The function variable values must correspond to the FUNCTION numbers, i.e., 1 for FUNC 1, 2 for FUNC 2, etc.
FNUMBER is 3
FNUM 3
FNUMBER should be specified prior to DATA.
FUNCTION model block: The FUNCTION-END block contains statements to define the function to be fit to the data. The argument N indicates the function number.
FUNC 1
F=A*EXP(B)+C*EXP(D)
END
FUNC 1
F=A*EXP(B)+C*EXP(D)
END
FUNC 2
F=Z(1)
END
A function block must be used for every dataset to be fit. NFUNCTIONS specifies the number of function blocks. The variable F must be assigned the function value. WT can be assigned a value to use as the weight for the current observation.
INITIAL command: Specifies initial values for the estimated parameters. The arguments I1, I2, I3, …, IN are the initial values, separated by commas or spaces.
INITIAL=0.01, 5.e2, -2.0
INIT are 0.02, 10, 1.3e-5
INIT 0.1 .05 100
INITIAL must be preceded by NPARAMETERS.
ITERATIONS command: Specifies the maximum number of iterations to be executed, N.
ITERATIONS are 30
ITER 20
If ITERATIONS=0, the usual output is produced using the initial estimates as the final values of the parameters.
LOWER command: Specifies lower bounds for the estimated parameters. The arguments L1, L2, L3, …, LN are the lower bounds, separated by commas or spaces.
LOWER 0 0 10
If LOWER is used, a lower bound must be provided for each parameter, and UPPER must also be used. LOWER must be preceded by NPARAMETERS.
MEANSQUARE command: Allows a specified value other than the residual mean square to be used in the estimates of the variances and standard deviations. The argument M replaces the residual sum of squares in the estimates of variances and standard deviations. If given, MEANSQUARE must be greater than zero.
MEANSQUARE=100
MEAN=1.0
METHOD command: In modeling, the METHOD command specifies the type of fitting algorithm Phoenix is to use. The argument N takes the following values:
1 Nelder-Mead Simplex
2 Levenberg and Hartley modification of Gauss-Newton (default)
3 Hartley modification of Gauss-Newton
In noncompartmental analysis, METHOD specifies the method to compute area under the curve. The argument N takes on the following values:
1 Lin/Log-Linear trapezoidal rule up to Tmax, and log trapezoidal rule after
2 Linear trapezoidal rule (Linear interpolation) (default)
3 Linear up/Log down rule: Uses the linear trapezoidal rule any time the concentration data is increasing and the logarithmic trapezoidal rule any time that the concentration data is decreasing.
4 Linear Trapezoidal (Linear/log Interpolation)
METHOD is 2
METH 3
MODEL command: In a command file, MODEL specifies the compiled or source model to use.
If MODEL N is specified, model N is used from the built-in library. If MODEL N, PATH is used, model N from the source file path is used.
MODEL 5
MODEL 7, 'PK'
MODEL 3 'D:\MY.LIB'
MODEL LINK command: MODEL with the option LINK specifies that compiled models are to be used to perform PK/PD or Indirect Response modeling. M1 is the PK model number, and M2 is the model number of the PD model or Indirect Response model. When this option is used a PKVAL command must also be used.
MODEL LINK 4 101
PKVAL 10 1.5 1.0 1.0
MODE LINK 1, 53
PKVA 10, 2.5
See also PKVALUE.
NCARRY command: Specifies the number of carry-over variables specified. The argument N is the number of carry-over variables.
NCAR are 3
NCAR 1
NCARRY must be specified before CARRYALONG.
NCATRANS command: When using NCA, NCATRANS specifies that the Final Parameters output will present each parameter value in a separate column, rather than row.
NCATRANS
NCAT
NCONSTANTS command: The NCONSTANTS command specifies the number of constants, N, required by the model. If used, NCONSTANTS must precede CONSTANTS.
NCONSTANTS are 3
NCON 1
NDERIVATIVES command: Indicates the number, N, of differential equations in the system being fit.
NDERIVATIVES are 2
NDER 5
NFUNCTIONS command: Indicates the number, N, of functions to be fit (default=1).
There must be data available for each function. If used, NFUNCTIONS must precede DATA.
NFUNCTIONS are 2
NFUN 3
The functions can contain the same parameters so that different datasets can contribute to the parameter estimates. When fitting multiple functions one must delineate which observations belong to which functions. To do this, include a function variable in the dataset or use NOBSERVATIONS. See FNUMBER or NOBSERVATIONS.
NOBOUNDS command: Tells Phoenix not to compute bounds during parameter estimation.
NOBOUNDS
NOBO
Unless a NOBOUNDS command is supplied Phoenix will either use bounds that the user has supplied, or compute bounds.
NOBSERVATIONS command: Provides backward compatibility with PCNonlin command files. When more than one function is fit simultaneously, NOBS can indicate which observations apply to each function. The arguments N1, N2,…, NF are the number of observations to use with each FUNCTION block. The data must appear in the order used: FUNC 1 data first, then FUNC 2, etc. Note that this statement requires counting the number of observations for each function, and updating NOBS if the number of observations changes.
NOBS must precede DATA.
NOBS 10, 15, 12
The recommended approach is to include a function variable whose values indicate the FUNCTION block to which the data belong. (See FNUMBER.)
NOPAGE command: Tells Phoenix to omit page breaks in the Core Output file.
NOPAGE
NOPAGE BREAKS ON OUTPUT
NOPLOTS command: Specifies that no plots are to be produced.
NOPLOTS
This command turns off both the high resolution plots and Core Output.
NOSCALE command: Turns off the automatic scaling of weights when WEIGHT is used. By default, the weights are scaled such that the sum of the weights equals the number of observations with non-zero weights.
NOSCALE
This command will have no effect unless WEIGHT is used.
NOSTRIP command: Turns off curve stripping for all profiles.
NOSTRIP
NPAUC command: Specifies the number of partial area under the curve computations requested.
NPAUC 3
See PAUC.
NPARAMETERS command: Sets the number of parameters, N, to be estimated. NPARAMETERS must precede INITIAL, LOWER or UPPER. (LOWER and UPPER are optional.)
NPARAMETERS 5
NPAR 3
See PNAMES.
NPOINTS command: Determines the number of points in the Predicted Data file. Use this option to create a dataset to plot a smooth curve. 10 <= N <= 20,000.
NPOINTS 200
For compiled models without differential equations the default value is 1000. If NDERIVATES> 0 (i.e., differential equations are used) this command is ignored and the number of points is equal to the number in the dataset. The default for user models is the number of data points. This value may be increased only if the model has only one independent variable.
NSECONDARY command: Specifies the number, N, of secondary parameters to be estimated.
NSECONDARY 2
NSEC=3
See SNAMES.
NVARIABLES command: Specifies the number of columns in the data grid. N is the number of columns (default=2). If used, NVARIABLES must precede DATA.
NVARIABLES 5
NVAR 3
P array: Contains the values of the estimated parameters for the current iteration. The index N specifies the Nth estimated parameter.
P(1)
T=P(3)-A
The values of the INITIAL command correspond to P(1), P(2), etc., respectively. NPARAMETERS specifies the value of N. PNAMES can be used to assign aliases for P(1), P(2), etc.
PAUC command: Lower, upper values for the partial AUC computations. If a specified time interval does not match times on the dataset, then Phoenix will generate concentrations corresponding to those times.
PAUC 0, 5, 17, 24
See NPAUC.
PKVALUE command: Used to assign parameter values to the PK model when using MODEL LINK.
MODEL LINK 4 101
PKVAL 10 1.5 1.0 1.0
MODE LINK 1, 53
PKVA 10, 2.5
PNAMES command: Specifies names associated with the estimated parameters. The arguments 'name1', 'name2', …,'nameN' are names of the parameters. Each may have up to eight letters, numbers, or underscores, and must begin with a letter. These names appear in the output and may be used in the modeling code.
PNAMES are 'ALPHA', 'BETA', 'GAMMA'
PNAM 'K01', 'K10', 'VOLU
The number of names specified must equal that for NPARAMETERS.
PUNITS command: Specifies the units associated with the estimated parameters. The arguments 'unit1', 'unit2', …,'unitN' are units and may be up to eight letters, numbers, operators or underscores in length. These units are used in the output.
PUNITS are 'hr', 'ng/mL', 'L/hr'
PUNI 'hr'
The number of units specified must equal that for NPARAMETERS.
REMARK command: Allows comments to be placed in a command file.
REMARK Y transformed to log(y)
REMA GAUSS-NEWTON
If a command takes no arguments or a fixed number of arguments, comments may be placed on the same line as a command after any required arguments.
REWEIGHT command: In the PK modeling module, REWEIGHT specifies that iteratively reweighted least squares estimates of the parameters are to be computed. The weights have the form WT=FW where W is the argument of REWEIGHT and F is the current value of the estimated functions. If W < 0 and F < 0 then the weight is set to zero.
REWEIGHT 2
REWE -.5
REWEIGHT is useful for computing maximum likelihood estimates of the estimated parameters.
The following statement in a model is equivalent to a REWEIGHT command: WT=exp(W log(F)), or WT=F**W. This command is not used with noncompartmental analysis.
S array: Contains the estimated secondary parameters. The index N specifies the Nth secondary parameter.
S(1)=-P(1)/P(2)
The range of N is set by NSECONDARY. Aliases may be assigned for S(1), S(2), etc. using SNAMES. The secondary parameters are defined in the SECONDARY section of the model.
SECONDARY model block: The secondary parameters are defined in the SECONDARY-END block. Assignments are made to the array S.
SECONDARY
S(1)=0.693/K10
END
Or, if the command SNAMES 'K10_HL' and a PNAMES command with the alias K10 have been included:
SECONDARY
K10_HL=0.693/K10
END
NSECONDARY must be used to set the number of secondary parameters.
SIMULATE command: Causes the model to be estimated using only the time values in the dataset and the initial values. It must precede the DATA statement, if one is included.
SIMULATE
SIMULATE may be placed in any set of Phoenix commands. When it is encountered, no data fitting is performed, only the estimated parameters and predicted values from the initial estimates are computed. All other output such as plots and secondary parameters are also produced.
SIZE command: N is the model size. Model size sets default memory requirements and may range from 1 to 6. The default value is 2.
SIZE 3
Applicable only when using ASCII models.
SNAMES command: Specifies the names associated with the secondary parameters. The arguments 'name1', 'name2', …, 'nameN' are names of the secondary parameters, aliases for S(1), S(2), etc. Each may be up to eight letters, numbers or underscores in length, and must begin with a letter. They appear in the output and may also be used in the programming statements.
SNAMES are 'AUC', 'Cmax', 'Tmax'
SNAM 'K10_HL' 'K01_HL'
SORT command: Causes Phoenix to segment a large data file into smaller subsets. Each subset will be run through the modeling separately. Arguments s1 and the options s2, s3, etc. are the column numbers of the sort keys. The 1 that follows the command SORT has no function. It is included to provide compatibility with PCNonlin version 4.
SORT 1, 1
SORT 1 3 2
SORT 1, 1, 2
SORT must be on one line, i.e., it can not use the line continuation symbol '&.’
SPARSE command: Causes Phoenix to use sparse data methods for noncompartmental analysis. The default is to not use sparse methods. See “Sparse sampling calculation”.
STARTING model block: Statements in the STARTING-END block specify the starting values of the differential equations. Values are placed in the Z array. Parameters can be used as starting values.
START
Z(1)=CON(1)
Z(2)=0
END
STEADY STATE command: Informs Phoenix that data were obtained at steady state, so that different PK parameters are computed. # is Tau, the (assumed equal) dosing interval.
STEADY STATE 12
STEA 12
SUNITS command: Specifies the units associated with the secondary parameters. The arguments 'unit1', 'unit2', …,'unitN' are units and may be up to eight letters, numbers, operators or underscores in length. These units are used in the output.
SUNITS are 'ml', 'mL/hr'
SUNI '1/hr'
The number of units specified must equal NSECONDARY.
TEMPORARY model block: Statements placed on the TEMPORARY-END block define global temporary variables that can be used in the MODEL statements.
TEMP
T=X
DOSE1=CON(1)
TI=CON(2)
K21=(A*BETA+B*ALPHA)/(A+B)
K10=ALPHA*BETA/K21
K12=ALPHA+BETA-K21-K10
END
THRESHOLD command: Sets the (optional) threshold value for use analyzing effect data with NCA model 220. See “Drug effect data model 220” for details.
THRE 0
TIME command: Places the current time in the upper right hand corner of each output page.
TIME
TITLE command: Indicates that the following line contains a title to be printed on each page of the output. Up to five titles are allowed per run. To specify any title other than the first, a value N must be included with TITLE. 1 <= N <= 5.
TITLE
Experiment EXP-4115
TRANSFORM model block: Statements placed in the TRANSFORM-END block are executed for each observation before the estimation begins.
TRANSFORM
Y=exp(Y)
END
UPPER command: Sets upper bounds for the estimated parameters. The arguments U1, U2, U3, …, UN are the bounds, separated by commas or spaces.
UPPER 1, 1, 200
If UPPER is used, an upper bound must be provided for each parameter, and LOWER must also be specified. UPPER must be preceded by NPARAMETERS.
URINE command: Used to specify variables when doing noncompartmental analysis for urine data (Models 210–212). The user must supply: the beginning time of the urine collection interval, the ending times of the urine collection interval, the volume of urine collected, and the concentration in the urine. The four keywords LOWER UPPER VOLUME, and CONCENTRATION identify the required data columns; C1–C4 give their column numbers.
URINE LOWER=C1 UPPER=C2 VOLUME=C3 CONC=C4
URINE CONC C1 VOLU C2 LOWE C3 UPPE C4
The keywords may appear in any order, and only the first four letters of each keyword is required.
Variables must be specified for all four keywords.
WEIGHT command: Activates weighted least squares computations. The argument W sets the weight value: YW. If W is not set, a column in the dataset provides weights—by default, the third column. Another column may be set using WTNUMBER.
WEIGHT Uses the values in Column 3 as the weights
WEIGHT -.5 Uses as the weight 1/sqrt(Y)
WEIGHT
WTNUM 7 Uses the values in Column 7 as the weights
The weights are scaled so that their sum = the number of observations with non-zero weights.
See also WTNUMBER, WT, REWEIGHT and NOSCALE.
WT variable: In PK modeling, WT contains the weight for the current observation.
WT=1/(F*F)
If the variable WT is included in a model, then any weighting indicated via the Variables and Weighting dialog will be ignored.
WTNUMBER command: Specifies which column of the dataset contains weights. Not used for NCA.
WTNUMBER 4
WTNU is 7
If WEIGHT is used and WTNUMBER is not, the default column for weights is 3.
X variable: Contains the current value of the independent variable.
F=exp(-X)
By default, when used in a command file, the values of X are taken from the first column of the dataset unless the XNUMBER command has been used. If the variable is raised to a negative power, enclose the power in parentheses or write the expression as a fraction. For example, X**–2 must be written as X**(–2) or 1/X**2.
XNUMBER command: Sets the column number, N, of the independent variable. If differential equations are being fit, XNUMBER indicates the column number that the differential equations are integrated with respect to. The default value is 1.
XNUMBER is 3
XNUM 1
XNUMBER should precede DATA.
Y command: Current value of the dependent variable.
Y=log(Y)
By default, Y is assumed to be in the 2nd column unless YNUMBER is used.
YNUMBER command: The YNUMBER command specifies the column number, N, of the dependent variable. When running Version 4 command files, the default value is 2.
YNUMBER is 4
YNUM 3
The YNUMBER command should precede the DATA command.
Z array: Contains the current values of the solutions of the differential equations. The index N specifies the Nth differential equation.
B=A*Z(2)
The maximum value of N is set by NDERIVATIVES.
