Defined as relative bioavailability, bioequivalence involves comparison between test and reference drug products, where the test and reference products can vary, depending upon the comparison to be performed. Although bioavailability and bioequivalence are closely related, bioequivalence comparisons rely on a criterion, a predetermined bioequivalence limit, and calculation of a interval for that criterion.
Use one of the following to add the Bioequivalence object to a Workflow:
Right-click menu for a Workflow object: New > NCA and Toolbox > Bioequivalence.
Main menu: Insert > NCA and Toolbox > Bioequivalence.
Right-click menu for a worksheet: Send To > NCA and Toolbox > Bioequivalence.
Note:To view the object in its own window, select it in the Object Browser and double-click it or press ENTER. All instructions for setting up and execution are the same whether the object is viewed in its own window or in Phoenix view.
This section contains information on the following topics:
The Bioequivalence model object is based on a mixed effects model. For more see “General linear mixed effects model”.
Use the Main Mappings panel to identify how input variables are used in a bioequivalence model. A separate analysis is performed for each profile, or unique level of soft key(s). Required input is highlighted orange in the interface.
•None: Data types mapped to this context are not included in any analysis or output.
•Sort: Categorical variable(s) identifying individual data profiles, such as subject ID and treatment in a crossover study. A separate analysis is done for each unique combination of sort variable values.
•Subject: The subjects in a dataset.
•Sequence: The order of drug administration.
•Period: The washout period, or the time period between two treatments needed for drug elimination. Only applicable in a crossover study.
•Formulation: The treatment and reference drug formulations used in a study.
•Dependent: The dependent variables, such as drug concentration, that provides the values used to fit the model.
•Classification: Classification variables or factors that are categorical independent variables, such as formulation, treatment, and gender.
•Regressors: Regressor variables or covariates that are continuous independent variables, such as gender or body weight. The regressor variable can also be used to weight the dataset.
Input data considerations
-
Missing data: For population and individual bioequivalence, the application assumes complete data for each subject. If a subject has a missing observation, that subject is not included in the analysis. If the data have many missing observations, consider imputing estimated values to produce complete records per subject. Phoenix does not impute missing values.
-
Variable name and data limits: See “Data limits and constraints”.
Note:Be sure to finalize column names in your input data before sending the data to the Bioequivalence object. Changing names after the object is set up can cause the execution to fail.
The Model tab allows users to specify the bioequivalence model.
-
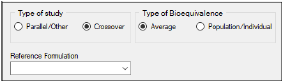
Under Type of study, select the Parallel/Other or Crossover option buttons to select the type of study.
-
Under Type of Bioequivalence, select the Average or Population/Individual option buttons to select the type of bioequivalence. Crossover studies are the only study type permitted when population or individual are the types of bioequivalence being determined.
-
In the Reference Formulation menu, select the reference formulation or treatment. The menu is only available after a formulation variable is mapped.
The Bioequivalence object automatically sets up default fixed effects, random effects, and repeated models for average bioequivalence studies depending on the type of study design: replicated crossover, nonreplicated crossover, or parallel. For more details on the default settings, see, see “Average bioequivalence study designs”.
For more information on population and individual bioequivalence, see “Population and individual bioequivalence”.
The Fixed Effects tab allows users to specify settings for study variables used in an average bioequivalence model. Population and individual bioequivalence models do not use fixed effects, so most options in the Fixed Effects tab are unavailable for population or individual bioequivalence models.
Average bioequivalence
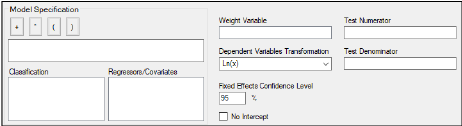
For average bioequivalence models the Model Specification field automatically displays an appropriate fixed effects model for the study type. Edit the model as needed.
Phoenix automatically specifies average bioequivalence models based on the study type selected and the dataset used. These default models are based on US FDA Guidance for Industry (January 2001).
See the following for details on the models used in a particular study type:
•Nonreplicated crossover designs
Study variables in the Classification box and the Regressors/Covariates box can be dragged to the Model Specification field to create the model structure.
-
Drag variables from the Classification and the Regressors/Covariates boxes to the Model Specification field and click the operator buttons to build the model:
 : addition
: addition  : multiplication
: multiplication  ,
,  : parentheses, for indicating nested variables in the model
: parentheses, for indicating nested variables in the model
Alternatively, the names and operators can be typed directly in the field.
Below are some guidelines for using parentheses:
•Parentheses in the model specification represent nesting of model terms.
•Seq+Subject(Seq)+Period+Form is a valid use of parentheses and indicates that Subject is nested within Seq.
•Drug+Disease+(Drug*Disease) is not a valid use of parentheses in the model specification.
-
Select a weight variable from the Regressors/Covariates box and drag it to the Weight Variable field.
To remove the weight variable, drag the variable from the Weight Variable field back to the Regressors/Covariates box.
The Regressors/Covariates box lists variables mapped to the Regressors context (in the Main Mappings panel). If a variable is used to weight the data then the variable is displayed in the Regressors/Covariates box. Below are some guidelines for using weight variables:
•The weights for each record must be included in a separate column in the dataset.
•Weight variables are used to compensate for observations having different variances.
•When a weight variable is specified, each row of data is multiplied by the square root of the corresponding weight.
•Weight variable values should be proportional to the reciprocals of the variances. Typically, the data are averages and weights are sample sizes associated with the averages.
•The Weight variable cannot be a classification variable. It must be declared as a regressor/covariate before it can be used as a weight variable. It can also be used in the model.
-
In the Dependent Variables Transformation menu, select one of the transformation options:
•None
•Ln(x): Linear transformation
•Log10(x): Logarithmic base 10 transformation
•Already Ln-transformed: Select if the dependent variable values are already transformed.
•Already Log10-transformed: Select if the dependent variable values are already transformed.
-
In the Fixed Effects Confidence Level box, type the level for the fixed effects model. The default value is 95%.
-
By default, the intercept term is included in the model (although it is not shown in the Model Specification field), check the No Intercept checkbox to remove the intercept term.
-
Use the Test Numerator and Test Denominator fields to specify an additional test of hypothesis in the case of a model with only fixed effects.
For this case, the default error term (denominator) is the residual error, so an alternate test can be requested by entering the fixed effects model terms to use for the numerator and denominator of the F-test. The terms entered must be in the fixed effects model and the random/repeated models must be empty for the test to be performed. (See “Tests of hypotheses” for additional information.) -
In the Dependent Variables Transformation menu, select Ln(x) or Already Ln-transformed.
•Ln(x): Linear transformation
•Already Ln-transformed: Select if the dependent variable values are already transformed.
The Variance Structure tab allows users to set random effects and repeated specification for the bioequivalence model. Users can also set traditional variance components and random coefficients. The Variance Structure tab is only available for average bioequivalence models.
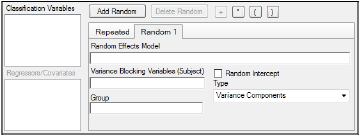
Users can specify none, one, or multiple random effects. The random effects specify Z and the corresponding elements of G=Var(g). Users can specify only one repeated effect. The repeated effect specifies the R=Var(e).
Phoenix automatically specifies random effects models and repeated specifications for average bioequivalence models. For more on the default models and specifications, see “Recommended models for average bioequivalence”.
The random effects model can be created using the classification variables.
•Use the pointer to drag the variables from the Classification Variables box to Random 1 tab.
•Users can also type variable names in the fields in the Random 1 tab.
The random effects model can be created using the regressor or covariate variables.
•Use the pointer to drag the variables from the Regressors/Covariates box to Random 1 tab.
•Users can also type variable names in the fields in the Random 1 tab.
The Random 1 tab is used to add random effects to the model. The random effects are built using the classification variables, the regressors/covariates variables, and the operator buttons.
The Repeated tab is used to specify the R matrix in the mixed model. If no repeated statement is specified, R is assumed to be equal to s2I. The repeated effect must contain only classification variables.
The default repeated model depends on whether the crossover study is replicated or not.
Caution:The same variable cannot be used in both the fixed effects specification and the random effects specification unless it is used differently, such as part of a product. The same term (single variables, products, or nested variables) must not appear in both specifications.
-
Drag variables from the boxes on the left to the fields in the tab and click the operator buttons to build the model.
 : addition, not available in the Repeated tab or when specifying the variance blocking or group variables
: addition, not available in the Repeated tab or when specifying the variance blocking or group variables  : multiplication
: multiplication  ,
,  : parentheses, for indicating nested variables in the model
: parentheses, for indicating nested variables in the model
Alternatively, type the names and operators directly in the fields. -
The Variance Blocking Variables (Subject) field is optional and, if specified, must be a classification model term built from the items in the Classification Variables box. This field is used to identify the subjects in a dataset. Complete independence is assumed among subjects, so the subject variable produces a block diagonal structure with identical blocks.
-
The Group field is also optional and, if specified, must be a classification model term built from items in the Classification Variables box. It defines an effect specifying heterogeneity in the covariance structure. All observations having the same level of the group effect have the same covariance parameters. Each new level of the group effect produces a new set of covariance parameters with the same structure as the original group.
-
(Random 1 tab only) Check the Random Intercept checkbox to set the intercept to random.
This setting is commonly used when a subject is specified in the Variance Blocking Variables (Subject) field. The default setting is no random intercept. -
If the model contains random effects, the covariance structure type must be specified from the Type menu. Covariance structure type options include:
•Variance Components
•Unstructured
•Banded Unstructured (b), type the number of bands in the Number of bands(b) field (default is 1)
•Compound Symmetry
•Heterogeneous Compound Symmetry
•Autoregressive
•Heterogeneous Autoregressive
•No-Diagonal Factor Analytic
•Banded No-Diagonal Factor Analytic (f), type the number of factors in the Number of factors (f) field (default is 1)
•Toeplitz
•Banded Toeplitz (b), type the number of bands in the Number of bands(b) field (default is 1)
The number of factors or bands corresponds to the dimension parameter. For some covariance structure types this is the number of bands and for others it is the number of factors. For explanations of covariance structure types, see “Covariance structure types”.
-
Click Add Random to add additional variance models.
-
Click Delete Random to delete a variance model.
Settings in the Options tab change depending on whether the model type is average or population/individual.
Bioequivalence Options tab
Average bioequivalence options
-
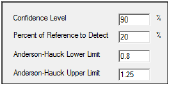
In the Confidence Level field, type the level for the bioequivalence model (default is 90%).
-
In the Percent of Reference to Detect field, type the percentage of reference treatment to detect (default is 20%).
-
In the Anderson-Hauck Lower Limit field, type the lower limit for the Anderson-Hauck test (default is 0.8).
-
In the Anderson-Hauck Upper Limit field, type the upper limit for the Anderson-Hauck test (default is 1.25 for log-transformed data and 1.2 for non-transformed data).
For more on the Anderson-Hauck test, see “Anderson-Hauck test”.
Population/Individual bioequivalence options
-
In the Confidence Level field, type the level for the bioequivalence model (default is 95%).
-
In the Percent of Reference to Detect field, type the percentage of reference treatment to detect (default is 20%).
-
In the Total SD Standard field in the Population Limits group, type the value of the total slope distance (default is 0.2).
-
In the Epsilon field in the Population Limits group, type an epsilon value for the population limits (default is 0.02).
-
In the Within Subject SD Standard field in the Individual Limits group, type the value of the within subject slope distance (default is 0.2).
-
In the Epsilon field in the Individual Limits group, type an epsilon value for the individual limits (default is 0.05).
The General Options tab is used to set output and calculation options for a bioequivalence model. The options change depending on whether the model is average bioequivalence or population/individual bioequivalence.
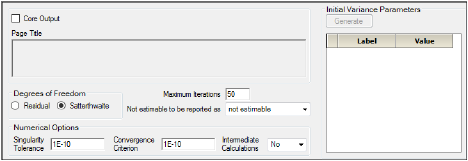
Bioequivalence General Options tab
Average bioequivalence options
-
Check the Core Output checkbox to include the Core Output text file in the results.
-
In the Page Title field, type a title for the Core Output text file.
-
Choose the degrees of freedom calculation method:
•Residual: The same as the calculation method used in a purely fixed effects model.
•Satterthwaite: The default setting and computes the df base on C2 approximation to distribution of variance.
-
In the Maximum Iterations field, type the number of maximum iterations. This is the number of iterations in the Newton fitting algorithm. The default setting is 50.
-
Use the Not estimable to be reported as menu to determine how output that is not estimable is represented.
•not estimable
•0 (zero)
-
In the Singularity Tolerance field, type the tolerance level. The columns in X and Z are eliminated if their norm is less than or equal to this number. (Default tolerance value is 1E–10.)
-
In the Convergence Criterion field, type the criterion used to determine if the model has converged (default is 1E–10).
-
In the Intermediate Calculations menu, select whether (Yes) or not (No) to include the design matrix, reduced data matrix, asymptotic covariance matrix of variance parameters, Hessian, and final variance matrix in the Core Output text file.
-
In the Initial Variance Parameters group, click Generate to edit the initial variance parameters values.
Note:The Generate initial variance parameters option is available only if the model uses random effects.
-
Select a cell in the Value column and type a value for the parameter (default is 1).
If the values are not specified, then Phoenix uses the method of moments estimates.
To delete one or more of the parameters from the table:
•Highlight the row(s).
•Select Edit >> Delete from the menubar or press X in the main toolbar.
•Click the Selected Row(s) option button and click OK.
Population/Individual bioequivalence options
-
Select the Core Output checkbox to include the Core Output text file in the results.
-
In the Page Title field, type a title for the Core Output text file.
