Blocks are sets of statements grouped together inside curly brackets. Statements include assignment of values and variables. Statements within the model (and within blocks) can span multiple lines of code, and can be separated with (optional) semicolons.
In the following statement list:
var = a variable name
varlist = a list of variables, optionally separated by commas
numlist = a list of numbers, optionally separated by commas
assign = an assignment operator
expr = any numerical expression, including variable names, numbers and arithmetic operators
[ …] = the content of the brackets is optional
| = “or” and separates options
* = zero or more of an item can be displayed
+ = one or more of an item can be displayed
statement = a statement of the form: variable=expression
cfMacro(id,parameters, [strip = stripping_dose_covariate])
Macro-parameter model of concentration, using optional stripping dose. See “Closed-form models” for more information.
cfMacro(id,parameters, [first = abs_cpt_name])
Macro-parameter model of amount in central compartment. See “Closed-form models” for more information.
cfMicro(id,parameters, [first = abs_cpt_name])
Micro-parameter model. See “Closed-form models” for more information and also see “One-compartment first-order absorption, closed-form” for an example.
count(var,expr[,action])
Defines an occurrence count. See “Count statement for Count models” for details.
covariate((var)*)
Defines one or more variables as covariates. Include an empty pair of parentheses after the covariate name to indicate that it is categorical.
covariate(dose,time,Gender())
See “Covariates” for more information.
delay(expression, MeanDelayTime[ , shape = ShapeExpression]
[ , hist = HistExpression][ , dist = NameofDistribution])
Models delayed outcomes using either discrete delay or distributed delay (which involves convolution of the signal to be delayed and the probability density function of the delay time, where gamma, Weibull, and inverse Gaussian distribution are supported).
Expression: Signal to be delayed.
MeanDelayTime: The value (positive) associated with the mean delay time.
shape = ShapeExpression: (Optional) If provided, the delay function is used to describe a distributed delay with ShapeExpression (positive) representing the shape parameter that defines the distribution of the delay time. Otherwise, it is used to describe a discrete delay.
hist = HistExpression: (Optional) Specify the value of the expression prior to time 0. If not provided, then 0 is assumed.
dist = NameOfDistribution: (Optional) Specify the name of the distribution for the delay time. If not provided, gamma distribution is assumed.
Any number of delay functions can be included in a model. The MeanDelayTime and shape parameters can be estimated. See “Discrete and distributed delays” for more information.
delayInfCpt(A, MeanDelayTime, ParamRelatedToShape
[ , in = inflow][ , out = outflow][ , dist = NameOfDistribution])
A statement related to a compartment (that can receive a dose through a dosepoint statement) with all of its input delayed (where the distribution of the delay time is specified by the dist option), including the rate of administered dose (if provided) and the inflow specified by the “in” option (if provided).
A: Name of the compartment.
MeanDelayTime: The value (positive) associated with the mean delay time.
ParamRelatedToShape: The value of a parameter related to the shape parameter of the specified distribution: if the dist = InverseGaussian, then ParamRelatedToShape=ShapeParameter, otherwise, ParamRelatedToShape=ShapeParameter–1.
out = outflow: (Optional) Represents the flow (either out of or into compartment A) that is not delayed.
in = inflow: (Optional) Represents the additional inflow that is delayed.
dist = NameOfDistribution: (Optional) Represents the name of the distribution for the delay time.
See “Discrete and distributed delays” for more information.
gammaDelay(expression, MeanDelayTime, shape = ShapeParam
[, hist = HistExpression], numODE = NumberOfODEUsed)
Defines a gamma distributed delay.
Expression: signal to be delayed.
MeanDelayTime: The value (positive) associated with the mean delay time.
shape = ShapeParam: The value of the shape parameter for the gamma distribution
hist = HistExpression: (Optional) The value of the expression prior to time 0. If not provided, 0 is assumed.
numODE = NumberOfODEUsed: The number of ODEs used to approximate the gamma distributed delay.
See “Discrete and distributed delays” for more information.
deriv(var = expr)
Defines an ordinary differential equation.
deriv(a=-a*Cl/V)
See “Differential equation models” for more information.
dosepoint| dosepoint2(id [, tlag = expr][ , duration = expr]
[ , rate = expr][ , bioavail = expr)
Defines dosing. Example:
dosepoint(a1)
deriv(a1=-a1*ke1)
c1=a1/v1
dosepoint(a2)
deriv(a2=-a2*ke2)
c2=a2/v2
See “Dosing” for more information.
error(var[(freeze)][ = std])
Defines a Gaussian error variable with its standard deviation optionally provided. Examples:
error(CEps = 0.1)
error(Ceps(freeze)
See also “Observe statement for Gaussian Residual models”.
event(var,expr)
Defines an unscheduled (e.g., adverse) event, where var is an occur variable, expr is its hazard.
event(occur,haz)
The occur variable can be:
0=did not occur
1=occurred at the given time
2=occurred at least once in the prior interval
–n=occurred n times in the prior interval
–999999=no information about the prior interval
See also “Event statement for Time-to-event models” for more information
fixef(var[(freeze)][enable=int)]
[=c([lowerbound],[ initialestimate],[ upperbound])]
Defines variable(s) as fixed effects parameters, optionally using (freeze) to fix parameter value during the estimation process, with optional initial estimate and bounds.
See “Fixed effects” for more information.
interpolate((id)(*))
For more on the interpolate statement, see “Covariates”.
LL(obsvar,expr)
Defines a log-likelihood model where obsvar is the observed variable and expr is its log-likelihood. See “LL statement for user-defined log-likelihood models” for more information.
multi(var,invlink(,expr)*[ ,action])
Defines an integer-valued categorical observation, where invlink is an inverse link function and the rest is a series of ascending offset expressions that serve as inputs to the inverse link function.
multi(Y,ilogit,-C*slope+intercept0, -C*slope+intercept1), …)
See “Multi statement for Categorical models” for more information.
observe(id1[(id2)] [=expr][ ,bql][ ,dobefore={ …}][ ,doafter={ …}])
Defines a residual error model for a continuous observed variable, where id1 is the observed variable, id2 is the independent variable, and expr is some function of the prediction and a Gaussian/normal error variable.
observe(cObs=cpred+eps1)
See “Observe statement for Gaussian Residual models” for more information.
ordinal(var,invlink,input,slope, (,intercept)*)
Variation on the multi statement.
ordinal(Y,ilogit,C,slope,intercept0,intercept1)
See “Ordinal statement for Ordinal Responses” for more information.
Tmax=peak(Cmax=C)
or Tmax=peak(Cmax=C, max)
or Tmax=peak(Cmax=C, max=(t < 6))
or Tmin=peak(Cmin=C, min)
or Tmin=peak(Cmin=C, min=(t >= 6))
where Cmax and Cmin are the internal variables and do not need defining.
Use in model code (not in a sequence block, doafter, or dobefore). Tmax and Tmin are variable names of user choice, and C is any user-written expression. Variables Tmax and Cmax are initially 0, and hold their value until after a peak is detected. After a peak is detected, variables Tmax and Cmax are set to the peak of a cubic spline fitted to the points around the peak, where the points are what is seen on the trajectory of the differential equations.
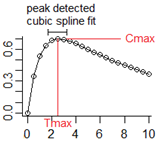
If a subsequent peak is found, the process will only be repeated if the peak is higher than the first. If the keyword max is not included, it is assumed. If the keyword min is included, it will look for a trough rather than a peak. Either keyword, max or min may be followed by a logical expression, such as max=test. In that case, it test is true, it looks for a peak. If test is false, it looks for the opposite.
peakreset(Cmax)
Use in a sequence block, doafter, or dobefore. This statement takes place at a point in time and will reset the peak-finder, after which it can detect another peak.
proc{ statement*}
ranef((diag|block(varlist) = numlist)*)
Defines variable(s) as random effects parameters, optionally defining the covariance matrix.
ranef(nlKe=0.01,nlV=0.01)
ranef(eta1,eta2=6
diag(eta5,eta6)=c(1,3)
same(eta7,eta8)
block(eta11,eta12)=c(1,2,3)
)
See “Random effects” for more information.
secondary((var = expr)+1)
Defines one or more secondary parameters, which are functions of fixed effects. For example, Tmax can be defined as a secondary parameter by using the built-in function CalcTMax as follows:
secondary(Tmax=CalcTMax(tvA,tvA1lpha,tvB,tvBeta,tvC,tvGamma))
section(<int>,section-stmt)
sequence{ statement*}
Defines a block of statements to be executed together and can contain conditional statements. It can contain special statements:
if statements set statements to be executed conditional on expr being true
if(expr){statement*}
[ else if(expr){statement*}]*
[ else {statement*}]
while statements set zero or more statements to be executed while expr is true
while (expr){statement*}
sleep statements pause computations for expr time units. (The expr value for the sleep statement is a relative time, not an absolute time.)
sleep (expr)
See “Modeling discontinuous events”, “Action code”, “The sequence and sleep statements”, “One-compartment model with sequence”, and “One-compartment model with sleep statement”.
stparm((var | var=expr) +)
Defines one or more structural model parameters, var, with optional values, expr.
stparm(
ka=exp(tvlKa+nlKa)
ke=exp(tvlKe+nlKe)
v=exp(tvlV+nlV)
)
See “Structural parameters and group statements” for more information.
urinecpt(var = expr)
Defines an elimination compartment. The urinecpt is like deriv, except in a steady state dosing situation, urinecpt is ignored.
