The Variance Structure tab allows users to set random effects and repeated specification for the bioequivalence model. Users can also set traditional variance components and random coefficients. The Variance Structure tab is only available for average bioequivalence models.
Users can specify none, one, or multiple random effects. The random effects specify Z and the corresponding elements of G=Var(g). Users can specify only one repeated effect. The repeated effect specifies the R=Var(e).
Phoenix automatically specifies random effects models and repeated specifications for average bioequivalence models. For more on the default models and specifications, see “Recommended models for average bioequivalence”.
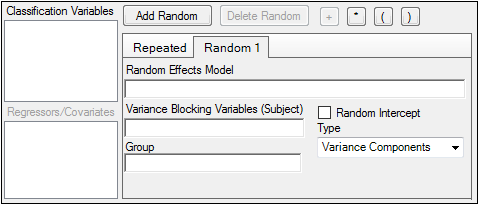
The random effects model can be created using the classification variables.
•Use the pointer to drag the variables from the Classification Variables box to Random 1 tab.
•Users can also type variable names in the fields in the Random 1 tab.
The random effects model can be created using the regressor or covariate variables.
•Use the pointer to drag the variables from the Regressors/Covariates box to Random 1 tab.
•Users can also type variable names in the fields in the Random 1 tab.
The Random 1 tab is used to add random effects to the model. The random effects are built using the classification variables, the regressors/covariates variables, and the operator buttons.
The Repeated tab is used to specify the R matrix in the mixed model. If no repeated statement is specified, R is assumed to be equal to s2I. The repeated effect must contain only classification variables.
The default repeated model depends on whether the crossover study is replicated or not.
Caution:The same variable cannot be used in both the fixed effects specification and the random effects specification unless it is used differently, such as part of a product. The same term (single variables, products, or nested variables) must not appear in both specifications.
-
Drag variables from the boxes on the left to the fields in the tab and click the operator buttons to build the model or type the names and operators directly in the fields.
+ addition (not available in the Repeated tab or when specifying the variance blocking or group variables),
* multiplication,
( ) parentheses for indicating nested variables in the model. -
The Variance Blocking Variables (Subject) field is optional and, if specified, must be a classification model term built from the items in the Classification Variables box. This field is used to identify the subjects in a dataset. Complete independence is assumed among subjects, so the subject variable produces a block diagonal structure with identical blocks.
-
The Group field is also optional and, if specified, must be a classification model term built from items in the Classification Variables box. It defines an effect specifying heterogeneity in the covariance structure. All observations having the same level of the group effect have the same covariance parameters. Each new level of the group effect produces a new set of covariance parameters with the same structure as the original group.
-
(Random 1 tab only) Check the Random Intercept checkbox to set the intercept to random.
This setting is commonly used when a subject is specified in the Variance Blocking Variables (Subject) field. The default setting is no random intercept. -
If the model contains random effects, the covariance structure type must be specified from the Type menu.
If Banded Unstructured (b), Banded No-Diagonal Factor Analytic (f), or Banded Toeplitz (b) is selected, type the number of bands in the Number of bands(b) field (default is 1).
The number of factors or bands corresponds to the dimension parameter. For some covariance structure types this is the number of bands and for others it is the number of factors. For explanations of covariance structure types, see “Covariance structure types”.
-
Click Add Random to add additional variance models.
-
Click Delete Random to delete a variance model.
