Right-click the selected model and choose Translate model from the menu.
Or
Click ![]() in the toolbar.
in the toolbar.
Select the format from the pop-up list:
To NONMEM
To R
To Matlab
To Berkely Madonna
Enter a name for the new file in the field (click ![]() to browse to a different location).
to browse to a different location).
Press Translate.
Pirana includes translators that will convert specific parts of NONMEM code to alternate NONMEM code.
ADVAN 1-6 to $DES: This option translates NM-TRAN models written in ADVAN routine (1-6) to ordinary differential equations (ODEs).
MU-referencing thetas-etas: Converts models written using normal- or log-normal hs, e.g.,
CL = THETA(1) * EXP(ETA(1))
will be converted into:
MU_1 = LOG(THETA(1)) ; ** MU-referenced by Pirana
CL = EXP(MU_1+ETA(1))
; Original equation: CL = THETA(1) * EXP(ETA(1))
$DES to difference equations: For some models written in ODEs, writing some parts of the model in difference equations can considerably reduce computational burden, while maintaining parameter precision (Petersson, K.J., et al. J Pharmacokinet Pharmacodyn. 2010 Oct. 37(5):493-506). Using this option translates all code written in $DES, other than the dA/dt system, to $PK and adds some required code (using MTIME).
The R translation options extract the parameter estimates for the structural model (q) and also the between subject variability matrix (W). A multi-dose simulation is automatically implemented (Pirana currently does not read in the dataset to extract dosing information). No residual error model is currently implemented in the R translators, but can be added by the user.
PKPDSim: This package is not available on CRAN, but may be sourced from GitHub, e.g., within R, use:
library(devtools)
install_github("ronkeizer/PKPDsim")
library(PKPDsim)
deSolve: The R code generated using this option is automatically loaded in the defined R interface.
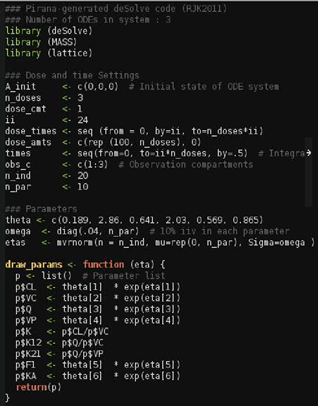
An example of generated R code is depicted in the following image.
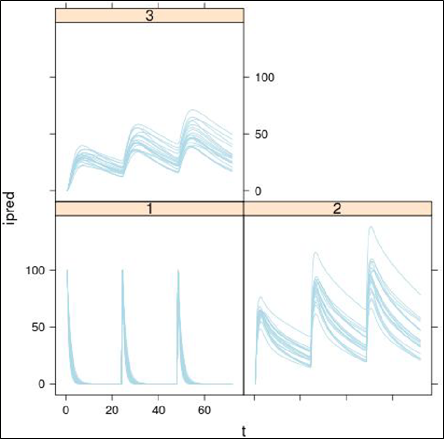
An example of associated simulation output for the deSolve-generated R code is depicted below.
The Matlab translation options extract the parameter estimates for the structural model (q) only (the between subject variability matrix is not extracted). No residual error model is currently implemented in the R translators, but can be added by the user.
to ODEs
to PopED files: Porting the model structure to PopED allows evaluation of optimal study designs (OD). This option creates the necessary files for PopED execution, however the details of the design and other optimization settings still need to be provided.
The generated Matlab code is automatically loaded in the defined code editor.
The Berkely Madonna translation option extracts the parameter estimates for the structural model (q) and also the between subject variability matrix (W). No residual error model is currently implemented in the R translators, but can be added by the user.
Check the Include between-subject variability box to consider variability between subjects. If checked, then choose the type of sampling to perform during the between-subject variability computations: Univariate sampling or Multivariate sampling.
The generated Berkeley Madonna code is automatically loaded in the defined code editor.
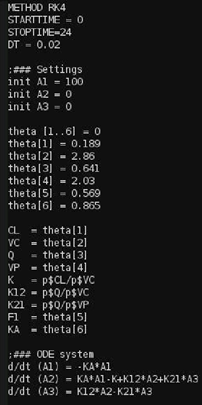
An example of generated Berkely Madonna code is depicted in the following image.