Automated Modeling Workflow Example
An automated modeling workflow for NONMEM models alleviates the burden on modeling scientist by removing the repetitive task of running and evaluating many candidate models, standardizes the model development between modelers, and standardize the results reported from such an analysis ultimately leading to higher quality of PopPK analyses (Schmidt et al. JPKPD 2014 Aug). In Pirana (version >= 2.10), such a workflow is made available, and in this tutorial we will walk through an example of an automated population PK analysis.
For this tutorial, we will use the template model library that is provided with Pirana, and a (simulated) dataset of an iv-administered drug also provided with Pirana (demo.csv).
Create a new project folder.
Browse into this folder (with Pirana) and copy the file demo.csv that is included in the Pirana installation folder (Pirana/automod_library/demo/demo.csv).
In Pirana, select the Tools > Automated modeling workflow > New analysis menu option.
Setting initial parameter estimates
The Step 1 dialog shows all models available in the library that can be selected for inclusion in the analysis. Use the filter field for conveniently selecting, e.g., only iv or only oral models. The dataset should of course be specified as well before advancing to the next step.
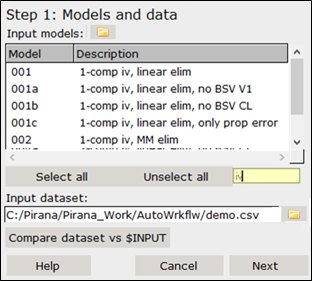
For this analysis, select all iv models by entering iv in the filter field of the Step 1 dialog.
Press the Ctrl key and select models 001, 002, 004, and 005 in the list.
When models and dataset have been selected, you should check whether the $INPUT record in the models matches with the headers in the dataset.
Click Compare dataset vs $INPUT. This will bring up the Compare headers dialog.
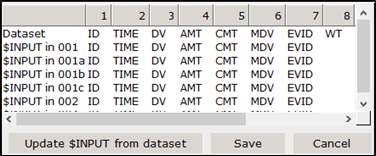
If the $INPUT in the models (shown in rows 2-…) does not match up with the dataset (shown in row 1), you can click the button Update $INPUT from dataset. This will create a new $INPUT record for all models. After clicking Save, when the models will be written (in step 3 of the automated analysis), the $INPUT records in all models will be changed to the new one. It is left to the user to make sure that the variables used in the model are still included in $INPUT, as there is no extra check in place for that.
Click Update $INPUT from dataset to update the records.
Click Save and then Next to advance to the next step.
Setting initial parameter estimates
In the Step 2 dialog, set initial parameter estimates, as well as lower and upper bounds. All parameters are read from the models that were selected in the Step 1 dialog. The parameter descriptions are defined in the models as comments to $THETA, $OMEGA, and $SIGMA blocks, e.g.,
$THETA
(0, 5, 100); CL
(0, 5, 100); V
$OMEGA
(0.1); CL
(0.1); V
$SIGMA
0.05 ; proportional error
Note: Correlations in $OMEGA and $SIGMA cannot be specified for an automated analysis. That is, only the diagonal elements of $OMEGA and $SIGMA can be specified in the template models if you want to update them in this step. You can still include models that have full $OMEGA or $SIGMA blocks as template model, however you cannot provide descriptions (as comments) to the parameters in the block, and you cannot update them in this step of the analysis.
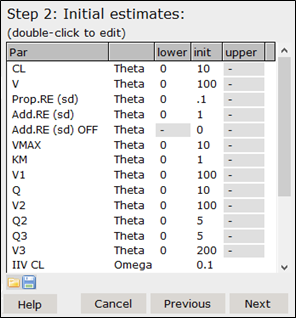
The two icons below the parameter list can be used to save (![]() ) parameter definitions to a csv file and load (
) parameter definitions to a csv file and load (![]() ) parameters from a csv file.
) parameters from a csv file.
For this analysis, leave the parameters as they are. Click Next to advance to the next step.
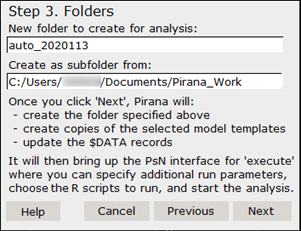
Use the Step 3 dialog to specify where to create the new models and run the analysis. By default, it will generate a new folder with a name based on the current date as a subfolder in the current folder in Pirana. This screen also lists the actions that Pirana will perform.
Use the defaults and click Next.
Pirana switches automatically to the new folder, where you will see the newly generated models. Pirana will also automatically bring up the PsN Toolkit (execute) dialog.
In this dialog, if you check the Advanced view box, you can select which R script(s) to run after all runs have been completed to generate goodness-of-fit plots.
Click ![]() next to the R scripts fields to select R scripts (or batch files) to run after (or before) the analysis step.
next to the R scripts fields to select R scripts (or batch files) to run after (or before) the analysis step.
For this analysis, select the NONMEM > Basic_GOF plots as single document to create GOF plots for all models.
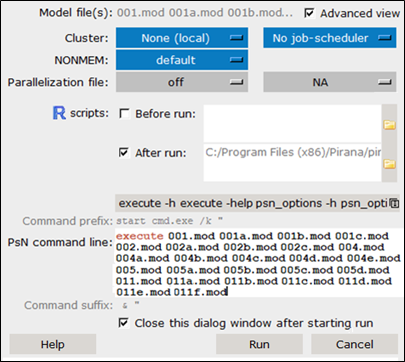
Press Run to execute the workflow.
The graphical report will automatically be opened, but is also available from the Reports tab on the right.
If you have not selected R scripts to be executed automatically after the analysis has completed, you can still create them afterwards by selecting the runs and running any R script from the R tab on the right side of the Pirana window.
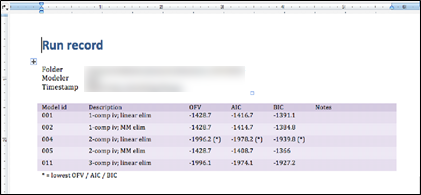
Besides the graphical report, Pirana can generate a numeric report for the analysis, including, e.g., OFVs, basic run information and parameter estimates. This document is not generated automatically but has to be requested manually after the analysis is complete:
Make sure Pirana is still in the folder where the analysis was run, and then go to Tools > Automated modeling workflow > Report.
On the first page you will see an overview of all models included in the analysis and their respective OFV, AIC and BIC. The subsequent pages includes information on each individual run in the analysis.