This example illustrates how a dataset can be fitted to a two-compartment model with first-order absorption in the pharmacokinetic model library using the Least-Squares Regression PK Model and then populate secondary parameters in a Maximum Likelihood model.
Note: The completed project (IndivPK.phxproj) is available for reference in …\Examples\NLME.
Set up the Least-Squares Regression PK model object
Create a new project called IndivPK.
Import the datasets IndivPK11.xls and IndivPK11_Dose.xls from …\Examples\NLME\Supporting files.
In the File Import Wizard dialog, press the arrow button to move from the IndivPK11 dataset to IndivPK11.
Press Finish.
Right-click the IndivPK11 worksheet and select Send To > Modeling > Least Squares Regression Models > PK Model.
Rename the object as WNL Model.
In the Main Mappings panel, map the columns to the contexts as follows:
Time to Time.
Conc to Concentration.
Select Dosing in the Setup tab.
Drag the IndivPK11_Dose worksheet from the Data folder to the Main Mappings panel.
Data types should automatically map as follows:
Time to Time.
Dose to Dose.
In the Model Selection tab, scroll down the table of models and check the box for model 11.
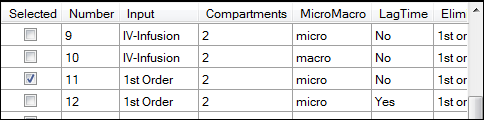
Select the Weighting/Dosing Options tab.
From Weighting menu select 1/Y.
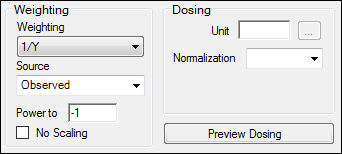
Click ![]() (Execute icon) to execute the object.
(Execute icon) to execute the object.
Create the Maximum Likelihood model object
Right-click the IndivPK11 worksheet and select Send To > Modeling > Maximum Likelihood Models.
Rename the object as NLME Model.
In the Structure tab, uncheck the Population box.
Press the Set WNL Model button.
The contents of the Structure tab changes. The first of the two untitled menus allows selecting a PK model, and the second allows selecting a PD model.
From the first untitled menu, select Model 11 (11 2cp micro xvas).
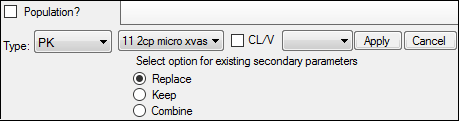
Since Model 11 is not a linked model, make sure the second untitled menu (where the linked model would be specified) is clear.
For this example, any secondary parameters that are already defined in the interface will be Replaced with those from the model.
Alternatively, Keep will ignore the model parameters and use the currently defined parameters. Combine keeps the existing parameters and adds those from the selected model to the list.
Press Apply to set the model.
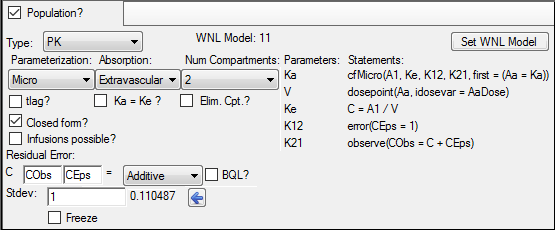
In the Main Mappings panel, map the columns to the contexts as follows:
Time to Time.
Conc to CObs.
Select Dosing in the Setup tab.
Drag the IndivPK11_Dose worksheet from the Data folder to the Main Mappings panel.
In the Dosing Mappings panel, map the columns to the contexts as follows:
Dose to Aa.
Time to Time.
Execute the object.
Initial Estimates configuration
The Initial Estimates tab can be used to find the best initial estimates for different parameters.
Select the Initial Estimates tab.
Adjust the portion of the plot to view by setting the Duration to 24 either using the slider or typing the value in the field next to the slider.
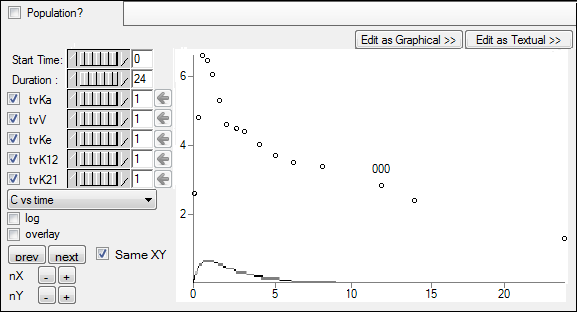
Enter different values for the fixed effect parameters, as shown in the table below, and see how each adjustment affects the results on the XY plot. Use the slider or typing the value in the field next to the slider.
tvKa = 2
tvV = 0.2 (make sure to re-check the box)
tvKe = 0.1 (make sure to re-check the box)
tvK12 = 1
tvK21 = 1
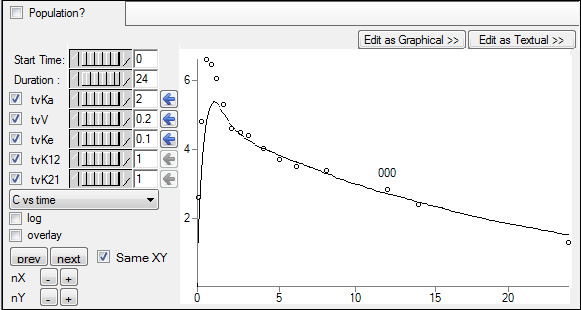
Click the left arrow button beside each parameter to copy the new estimate to the Initial estimate field.
For more information on using the Initial Estimates, see the “Initial Estimates tab” description.
Execute the object.
Comparing the results
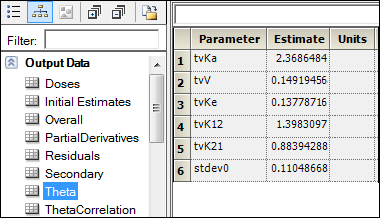
Open the Theta worksheet in the Results tab for the Phoenix Model object.
Open Final Parameters worksheet in the Results tab of the WNL Model object.
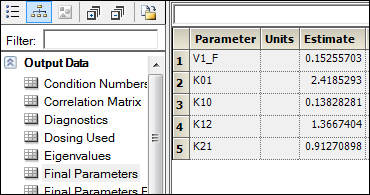
The names of parameters in Maximum Likelihood Models have the following equivalents in the PK Model.
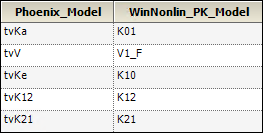
Compare the results of estimated fixed effects. As one can see, the estimates in these models are close but not equal due to different methods of model fitting.
This concludes the individual modeling example.