Analyzing variance structure effects
This analysis is concerned with the precision components of assay validation to estimate contributions due to assay variation, assay-to-assay variation, and analyst-to-analyst variation. A single QC sample was prepared containing, theoretically, 65 ng/mL of analyte. Five analysts were recruited for this study. Each analyst ran five aliquots of the sample on four assay runs. The data are available in the dataset AssayVal1.CSV in the Phoenix examples directory.
Note: The completed project (LinMix_StructureEffects.phxproj) is available for reference in …\Examples\WinNonlin.
Set up the object
Create a project called LinMix_StructureEffects.
Import the file …\Examples\WinNonlin\Supporting files\AssayVal1.CSV.
Units must be added to the Determination column before the dataset can be used in a Linear Mixed Effects model.
In the Columns tab below the table, select the Determination column header in the Columns list.
Clear the Unit field, type ng/mL, and press the Enter key.
Right-click Workflow in the Object Browser and select New > Computation Tools > Linear Mixed Effects.
Drag the AssayVal1 worksheet from the Data folder to the Main Mappings panel.
Map the columns to the contexts as follows:
Analyst to Classification.
Assay to Classification.
Determination to Dependent.
Select the Variance Structure tab below the Setup panel.
Drag Analyst from the Classification Variables list to the Random Effects Model field in the Random 1 sub-tab (or type Analyst in the field).
Press Add Random to add another Random effect.
Drag Assay from the Classification Variables list to the Random Effects Model field in the Random 2 sub-tab (or type Assay in the field).
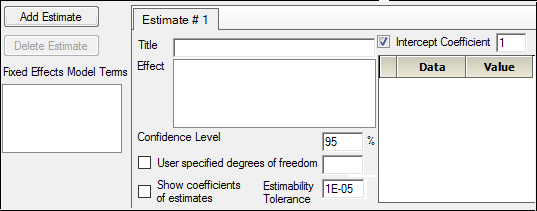
Select the Estimates tab.
Check the Intercept Coefficient box and type 1 in the Intercept Coefficient field.
Execute and view the results of the first model
Execute the object.
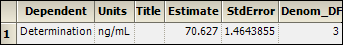
Select Estimates in the Results list.
Statistical accuracy values are located in the Estimates worksheet. The mean response is the intercept, which is estimated at 70.6 ng/mL with a 95% confidence interval. The lower interval is 65.97 and the upper interval is 75.28. Since the theoretical analyte concentration of 65 ng/mL is not within the interval, one can conclude that the bias is statistically significant. The method has a bias of approximately 5 ng/mL.

Select the Final Variance Parameters worksheet to view precision estimates and variance components.
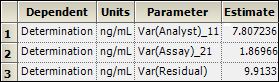
Based on these results, most of the variation is coming from analyst-to-analyst variation and from within-assay variation. Assay-to-assay noise is quite small. The units on the variances are (ng/mL).
Apply model to a different dataset
Now fit the same model to the data in the dataset AV3.CSV.
Import the file …\Examples\WinNonlin\Supporting files\AV3.CSV.
Units must be added to the Determination column before the dataset can be used in a Linear Mixed Effects model.
In the Columns tab, select the Determination column header in the Columns box.
Clear the Unit field, type ng/mL, and press the Enter key.
Right-click the Linear Mixed Effects object in the Object Browser and select Copy.
Right-click Workflow and select Paste.
A new Linear Mixed Effects object named Copy of Linear Mixed Effects is added to the Workflow. The LinMix object copy contains the same settings as the original object.
Map the AV3 dataset to Copy of Linear Mixed Effects. Do not change the data mappings in the Main Mappings panel.
Execute and view the results from the second model
Execute the object.
Select Final Variance Parameters in the Results list.
The Final Variance Parameters worksheet contains the following variance components:
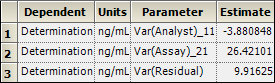
This table indicates that analyst-to-analyst variation is negative. Since variances cannot be negative, it is customary to replace the value with zero. A negative variance component indicates that the corresponding term should be removed from the model, which means that the contribution from that term is minimal compared to the contribution due to the other terms and it cannot be distinguished from the residual term.
From the variance components it is clear that the largest contribution to noise in the method is from run-to-run variation. Within-run variation also contributes to the noise. There is very little variation among analysts, indicating that the method is robust.
The Linear Mixed Effects object warns about negative final variances.
In the Results tab, select the Warnings and Errors text file.
The text file states: “Warning 11094: Negative final variance component. Consider omitting this VC structure.” Problems associated with a linear mixed effects model are written to this file during execution.
This concludes the LinMix structure effects analysis example.