Maximum Likelihood Model Comparer
The Maximum Likelihood Model Comparer is an operational object that can compare any executed Maximum Likelihood Model in a project, calculate differences between some model diagnostics, as well as calculate p-values for nested models.
Use one of the following to add the object to a Workflow:
Right-click menu for a Workflow object: New > Modeling > Maximum Likelihood Model Comparer.
Main menu: Insert > Modeling > Maximum Likelihood Model Comparer.
Right-click menu for a worksheet: Send To > Modeling > Maximum Likelihood Model Comparer.
Note:To view the object in its own window, select it in the Object Browser and double-click it or press ENTER. All instructions for setting up and execution are the same whether the object is viewed in its own window or in the Phoenix view.
This section contains information on the following topics:
User interface description
Results
The Setup tab lists all the Maximum Likelihood Models objects in a project.
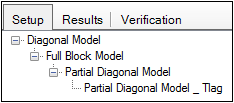
Note:Avoid having multiple Maximum Likelihood Models objects with the same name, even if they are in different workflows. Model Comparer will include all models with the same name in the comparison, even if their Compare boxes are not checked (they will become checked at execution). If the names cannot be changed for some reason, be sure to use the Hide checkboxes next to the model objects that are not wanted for the comparison.
If a Maximum Likelihood Models object was created by making a copy of a previous Maximum Likelihood Models object, then the copy is nested underneath the first model in the Comparison panel. The original model is the root model, and the copied model is the child.
Model copies that are nested underneath the original model will typically have additional parameters that are compared against the original model, which has fewer parameters.
In the Setup tab users can change the hierarchical relationship between root and sub-models.
-
Use the pointer to select a model in the Setup tab and drag the model on top of another model.
-
In the dialog, click Yes to confirm the move.
The selected model is nested underneath the selected root model.
-
To move a child model back to the root level, right-click the child model and select Orphan Model.
-
In the dialog, click Yes to confirm the move.
The model is moved back to the root level in the Setup tab.
Note:Although there are no icons in the Setup tab for importing/saving/loading object settings, these operations are still available using the File > Import menu, and right-clicking the object in the Object Browser or in the workflow diagram.
The area to the right of the model allows users to hide, remove, and select models to compare.
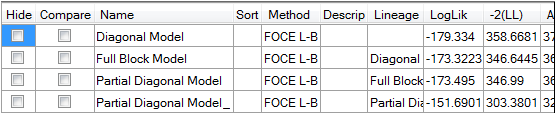
Check or uncheck the Hide or Compare checkboxes to exclude or include a model in the comparison.
•If the Hide checkbox is selected, then the model is considered “hidden” and is removed from any comparisons.
•If the Compare checkbox is selected, then the selected model is included in comparisons.
Additional model descriptions can be entered in the Description field by clicking the field twice and typing in the field. Description is carried over to the result worksheets.
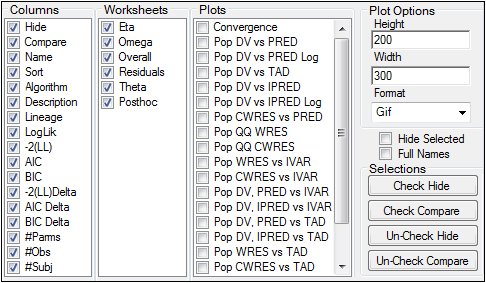
The Options tab lists all the columns, worksheets and plots available for comparisons.
-
Uncheck a column checkbox to remove that column from the Comparison panel. Removed columns are not included in the output worksheets.
-
Check the checkboxes in the Worksheets and Plots lists to include worksheets and plots in the comparison results.
-
Uncheck the checkboxes in the Worksheets and Plots lists to exclude the worksheets and plots from the comparison results.
-
In the Plot Options area, select the height, width, and graphic format for the output plot.
-
If a Maximum Likelihood Model object is marked as hidden, then check the Hide Selected checkbox to remove the model from the Setup tab. The hidden model is not used in any comparisons.
-
Check the Full Names checkbox to display the full name of a model. The full name includes the name of the workflow containing the model.
Use the buttons in the Selections area to select or hide several models at once. Multiple rows in the Comparison panel must be selected to use these buttons.
-
To select multiple rows in the Comparison panel, press and hold the CTRL key and right-click beside each row to select it.
Or
Use the pointer to select a row. Press and hold the SHIFT key and use the up or down arrow keys to select multiple rows. -
When the desired rows are selected, users can do the following:
Click Check Hide to hide the selected models.
Click Check Compare to include the selected models in the comparison.
Click Un-Check Hide to not hide the models.
Click Un-Check Compare to remove the models from the comparison.
Worksheet
-
Eta: Empirical Bayesian estimates of random differences from population expected values for each subject and occasion. This worksheet presents eta estimates by scenario and by model being compared.
-
Omega: Estimates of the random effects variance-covariance matrix, correlation matrix and shrinkage. This worksheet is presented by scenario and by model being compared.
-
Overall: Stacks each model 'Overall' worksheet result but doesn't present any comparison calculation. Columns include:
Name: Model names being compared (as displayed in the Object Browser)
Description: Model description text if entered by the user in the setup tab.
Scenario: Name of the scenario (if applicable)
Retcode: Code indicating the status of the run convergence
LogLik: Loglikelihood
-2LL: –2 Loglikelihood
AIC: Akaike Information Criterion for each model run
BIC: Bayesian Information Criterion for each model run
nParm: Number of model parameters
nObs: Number of observations
nSub: Number of subjects
EpsShrinkage: Epsilon shrinkage
-
Posthoc: Compares the Posthoc worksheet estimates of selected models. Model name, any description, scenario, replicate, ID, time, and one column for each structural parameter are listed.
-
Residuals: This table stacks 'Residuals' worksheet results of each individual model with the addition of Model name and Description. It contains columns for:
Name: Model name
Description: Model description
Scenario: Scenario name if applicable
ID: Subject ID
Time: Time
TAD: Time after dose
PRED: Population prediction
IPRED: Individual prediction
DV: Observed dependent variable. This column header will display the name of the column mapped to CObs for each individual run. If models being compared have different column names mapped to CObs then as many columns as CObs names used will display.
IRES: Individual residual
PREDSE: Standard error of predicted value
Weight: Applied model weight
IWRES: Individual weighted residual
WRES: Weighted residual
CWRES: Corrected weighted residual
PCWRES: Predictive check weighted residuals
CdfPCWRES: Cumulative distribution function for predictive check weighted residual. This column would only have non-zero values if the PCWRES option was selected in the individual runs.
CdfDV: Cumulative distribution function for the dependent variable. This column would only have non-zero values if the PCWRES option was selected in the individual runs.
TADSeq: Time after dose sequence. This variable is used to produce plots with TAD.
ObsName: Observation Name. This variable allows to distinguish observations when there are multiple observed quantities.
Var. Inf. factor: The variance inflation factor (if applicable)
-
Theta: This table stacks 'Theta' worksheet result of each individual model run with the addition of Model name and Description. It contains columns for:
Name: model name
Description: model description
Scenario: scenario name if applicable
Estimate: parameter estimates
Parameter: parameter name
Units: parameter units (if applicable)
Stderr: standard error
CV%: percent confidence of variation
lower % CI: lower confidence interval
upper % CI: upper confidence interval
Var. Inf. Factor: variance inflation factor (if applicable)
-
Comparison Results: List all columns checked under options. Columns may include:
Hide: Models hidden
Compare: Models to be compared in the plot table
Method: phoenix model engine used for each model
Name: is the name of the object being compared
Description: optional description of the model
Lineage: List the 'parent' (i.e. reduced model) if the current model is a 'child' (derived from but with additional parameters) of another model
LogLik: estimate of the loglikelihood upon convergence
AIC: Akaike Information Criterion for each model run
BIC: Bayesian Information Criterion for each model run
-2(LL)Delta: Difference in -2LL for nested models only (Lineage-Child model)
AICDelta: Difference in AIC values. for nested models only (Lineage-Child model)
BICDelta: Difference in BIC values for nested models only (Lineage-Child model)
#params: Number of model parameters
#obs: Number of observations
#Subj: Number of subjects
p-value: Chi-Square p-value based on the Likelihood Ratio Test for nested models only.
Plot
-
Plot Table: Illustration of all the selected plots in rows for each model being compared (columns). This plot table is not editable and it places side by side the plots from the individual runs for easy visualization. Any changes done to the individual run plots would reflect in this Plot table. This table can be exported or printed upon right click.
Text File
-
Settings: A text format file with the list of current columns, plots and models being compared.
